Capnodiales Woron.
MycoBank number: MB 90464; Index Fungorum number: IF 90464; Facesoffungi number: FoF 07632
Capnodiales was introduced by Woronichin (1925) and accommodated the families Antennulariellaceae, Capnodiaceae, Cladosporiaceae, Coccoideaceae, Dissoconiaceae, Metacapnodiaceae, Mycosphaerellaceae, Piedraiaceae and Teratosphaeriaceae (Lumbsch & Huhndorf 2010, Hyde et al. 2013, Chomnunti et al. 2011, 2014). Subsequently, Aeminiaceae (halotolerant on deteriorated limestones), Cystocoleaceae (lichenized), Euantennariaceae (plant parasitic), Extremaceae (extremophilic fungi), Johansoniaceae (epiphytic), Neodevriesiaceae (extremophilic fungi), Paradevriesiaceae (plant and rock-habitating fungi) surfaces, Phaeothecaceae (variety of life styles), Phaeothecoidiellaceae (sooty-blotch/flyspeck fungi), Racodiaceae (rock-habitating fungi), Schizothyriaceae (sooty-blotch/flyspeck fungi), and Xenodevriesiaceae (pathogenic or saprobic) were also accepted in this order based on phylogenetic analyses (Phookamsak et al. 2016, Hongsanan et al. 2017, Wijayawardene et al. 2017a, Doilom et al. 2018, Crous et al. 2019b), although type material of Schizothyriaceae needs to be recollected to stabilize this family. Paradevriesiaceae is synonymized under Extremaceae in this study base on its phylogenetic placement (Fig. 3). We provide a phylogenetic tree for Capnodiales (Fig. 3) including all families in this order. Phylogenetic trees of Mycosphaerellaceae and Teratosphaeriaceae are provided separately in Figs. 15 and 24.
Members of Capnodiales are mostly leaf epiphytes associated with honey dew (produced by insects), or saprobes, parasites and endophytes of plants worldwide. There have been several publications concerning rock-inhabiting fungi in the Capnodiales clade with phylogenetic analyses, but the connections between rock-inhabiting fungi and other lifestyles (i.e. plant pathogens and saprobes) found in this order are unexplained. Hongsanan et al. (2016a) provided the MCC tree for a better understanding of evolution of capnodialean families. The MCC tree answers ecological and evolutionary questions, concerning the adaptation of these groups to extreme environments. The common ancestor of species occurring in extreme habitats, such as species in Extremaceae and Neodevriesiaceae diverged after other families in Capnodiales, while earlier diverged families mostly comprise pathogens and saprobes (Ismail et al. 2016, Hongsanan et al. 2016a). In our analyses, the divergence time for Capnodiales is estimated as 221 MYA (stem age) (Fig. 2).
Accepted families: Aeminiaceae, Antennulariellaceae, Capnodiaceae, Cladosporiaceae, Cystocoleaceae, Dissoconiaceae, Euantennariaceae, Extremaceae, Johansoniaceae, Metacapnodiaceae, Mycosphaerellaceae, Neodevriesiaceae, Phaeothecaceae, Phaeothecoidiellaceae, Piedraiaceae, Racodiaceae, Schizothyriaceae, Teratosphaeriaceae and Xenodevriesiaceae.
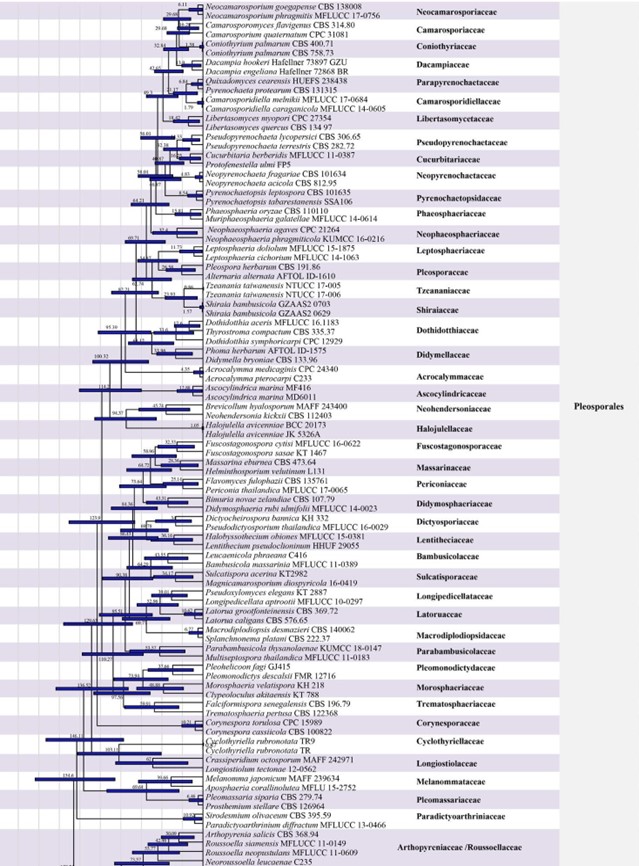
Figure 2 – The maximum clade credibility (MCC) tree of families in Dothideomycetes obtained from a Bayesian approach (BEAST). The fossil minimum age constraints and second calibrations used in this study are marked with green dots. Bars correspond to the 95 % highest posterior density (HPD) intervals. The scale axis shows divergence times as millions of years ago (MYA). Geological periods are indicated at the base of the tree.
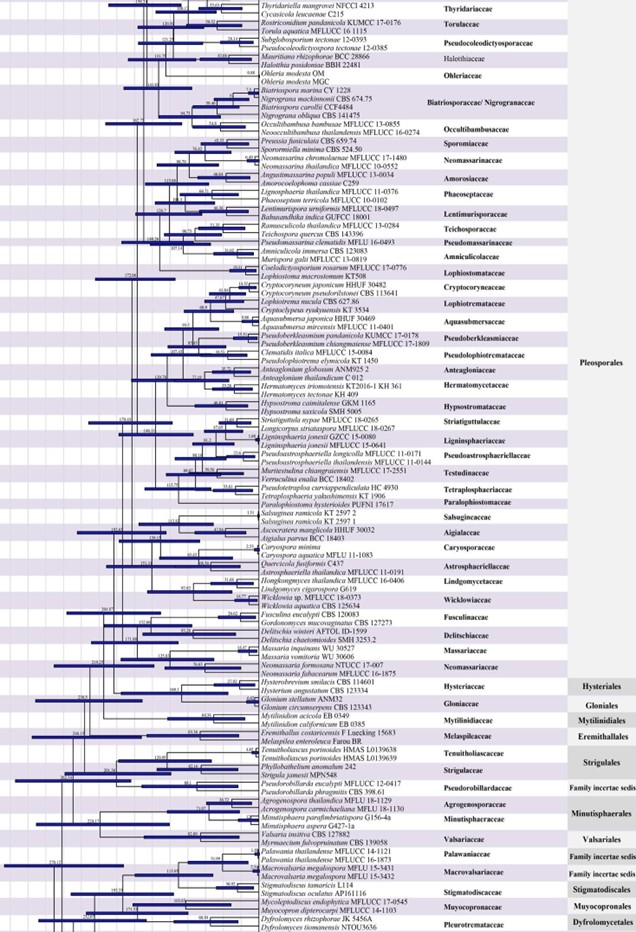
Figure 2 – Continued.
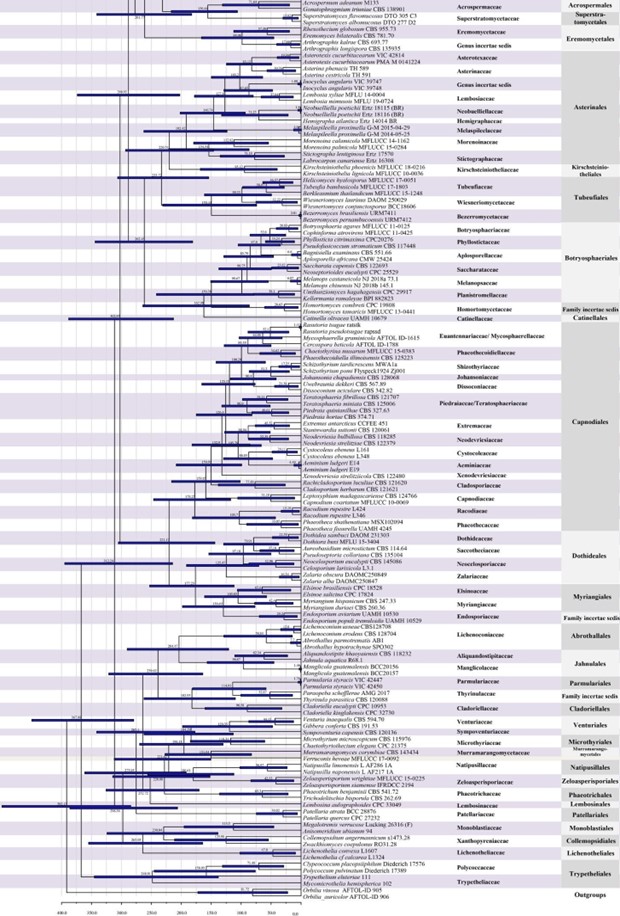
Figure 2 – Continued.
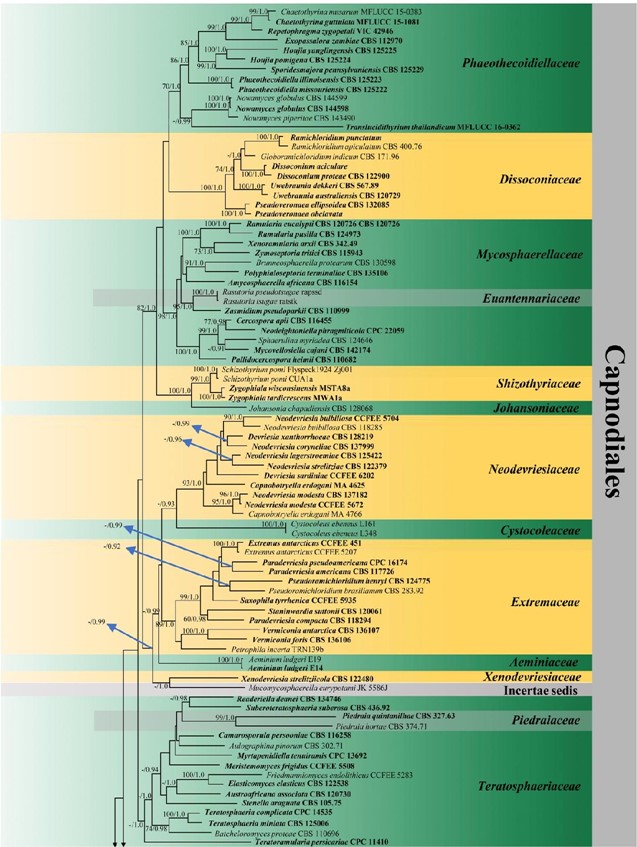
Figure 3 – Phylogram generated from maximum likelihood analysis (RAxML) of Capnodiales based on ITS, LSU and rpb-2 sequence data. Maximum likelihood bootstrap values equal or above 70 %, Bayesian posterior probabilities equal or above 0.90 (MLBS/PP) are given at the nodes. An original isolate number is noted after the species name. The tree is rooted to Elsinoe phaseoli (CBS 165.31). The ex-type strains are indicated in bold. Hyphen (-) represents support values below 70 % MLBS and 0.90 PP.
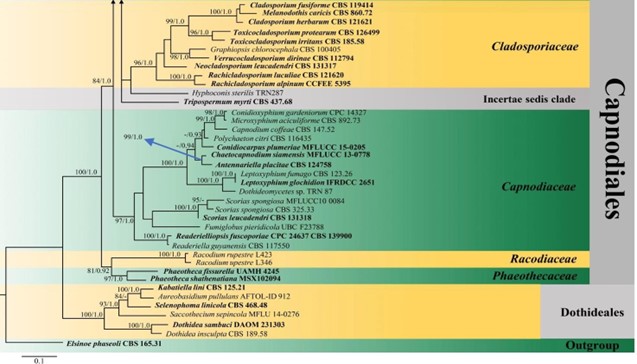
Figure 3 – Continued.
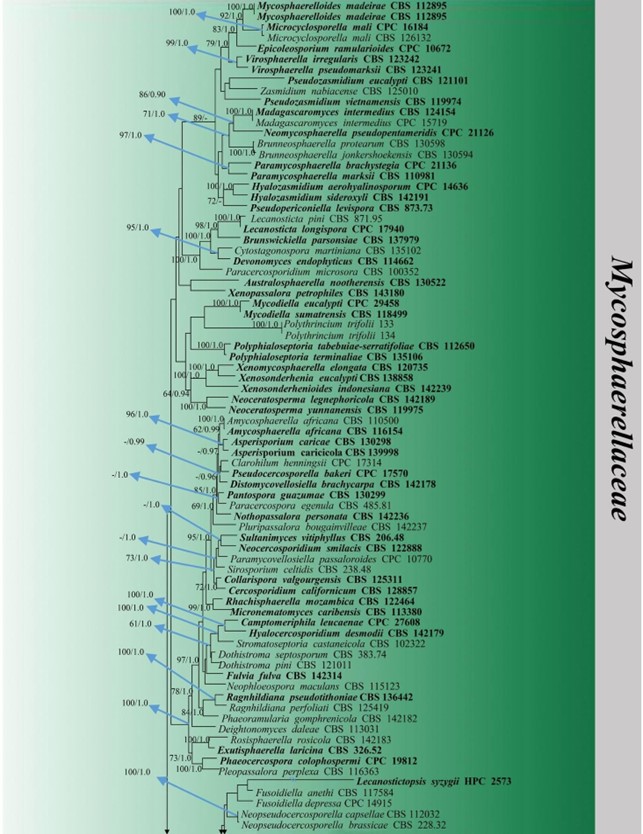
Figure 15 – Phylogram generated from maximum likelihood analysis (RAxML) of Mycosphaerellaceae based on ITS, LSU and rpb-2 sequence data. Maximum likelihood bootstrap values equal or above 70 %, Bayesian posterior probabilities equal or above 0.90 (MLBS/PP) are given at the nodes. An original isolate number is noted after the species name. The tree is rooted to Phaeotheca fissurella (UAMH 4245). The ex-type strains are indicated in bold. Hyphen (-) represents support values below 70 % MLBS and 0.90 PP.
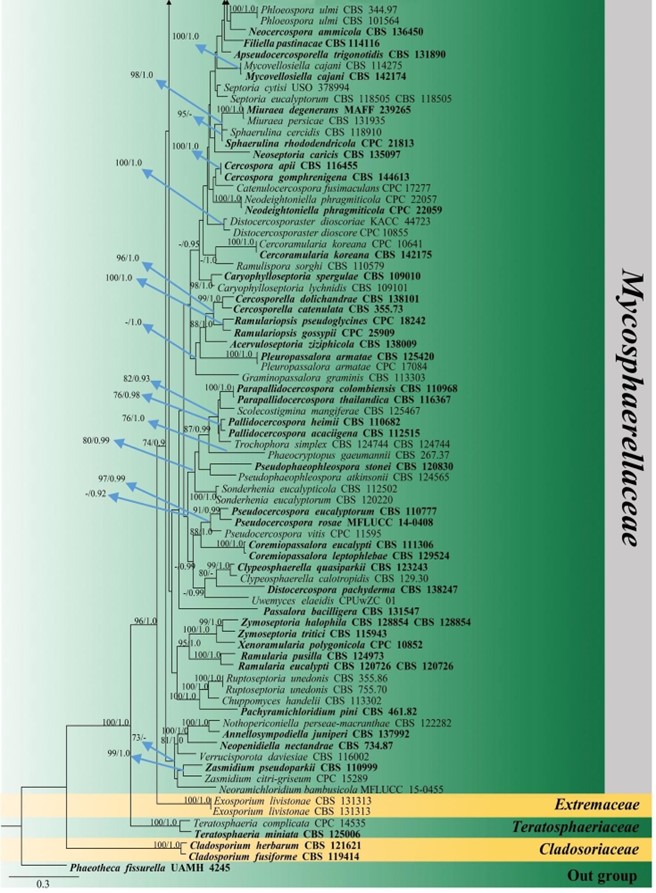
Figure 15 – Continued.
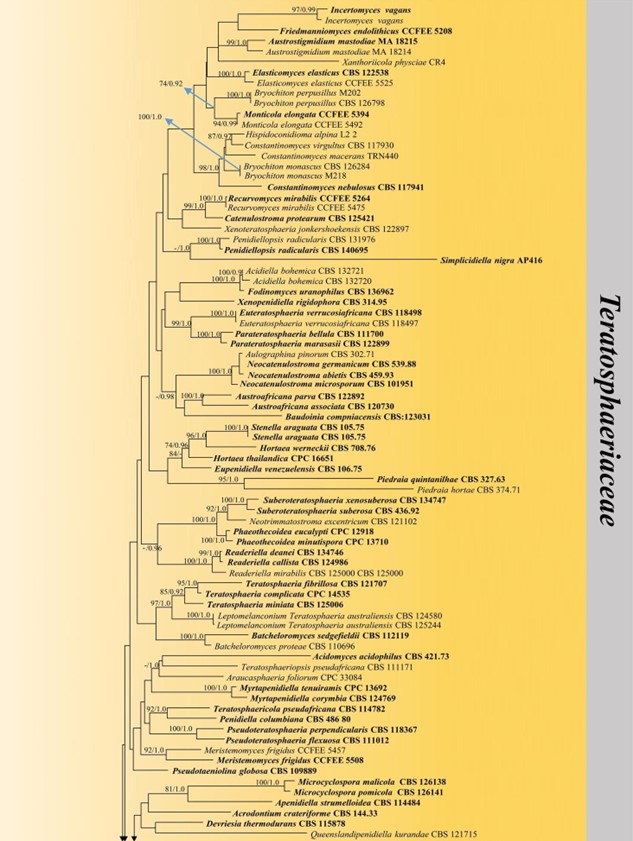
Figure 24 – Phylogram generated from maximum likelihood analysis (RAxML) of Teratosphaeriaceae based on ITS, LSU and rpb-2 sequence data. Maximum likelihood bootstrap values equal or above 70 %, Bayesian posterior probabilities equal or above 0.90 (MLBS/PP) are given at the nodes. An original isolate number is noted after the species name. The tree is rooted to Capnodium coartatum (MFLUCC 10-0069) and C. coffeae (CBS 147.52). The ex-type strains are indicated in bold. Hyphen (-) represents support values below 70 % MLBS and 0.90 PP.
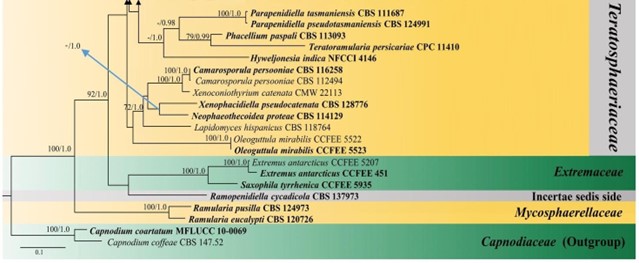
Figure 24 – Continued.
Families
Capnodiales genera incertae sedis
