Jobellisiales M.J. D’souza & K.D. Hyde, Fungal Divers. 72: 219 (2015)
MycoBank number: MB 551134; Index Fungorum number: IF 551134; Facesoffungi number: FoF 00621;
Jobellisiales was established by Maharachchikumbura et al. (2015) and comprises a single family Jobellisiaceae based on perithecial, yellow, orange or brown ascomata and cylindrical asci with brown ascospores (Ranghoo et al. 2001, Liu et al. 2012, Maharachchikumbura et al. 2016b). Most members in Jobellisiales have been found in America in terrestrial and freshwater habitats and no asexual morph is known (Ranghoo et al. 2001, Liu et al. 2012, Maharachchikumbura et al. 2016b). Jobellisiales species are similar to some members of Diaporthales in having brown, 1- septate ascospores (Maharachchikumbura et al. 2015, Senanayake et al. 2017a). Phylogenetically, Jobellisiales was a sister clade of Calosphaeriaceae (Maharachchikumbura et al. 2015, 2016b, Hongsanan et al. 2017), and Hyde et al. (2017a) proposed that Jobellisiales is closely related to Togniniaceae. Hongsanan et al. (2017) and Wijayawardene et al. (2018a) considered Jobellisiales to be synonym of Calosphaeriales. However, Jobellisiales is an unstable clade, and we accept it as an independent order and related to Calosphaeriales and Diaporthales (Fig. 12). The divergence time for Jobellisiales is estimated as 138 MYA (Fig. 2). Currently there is one family with one genus in this order (this paper).
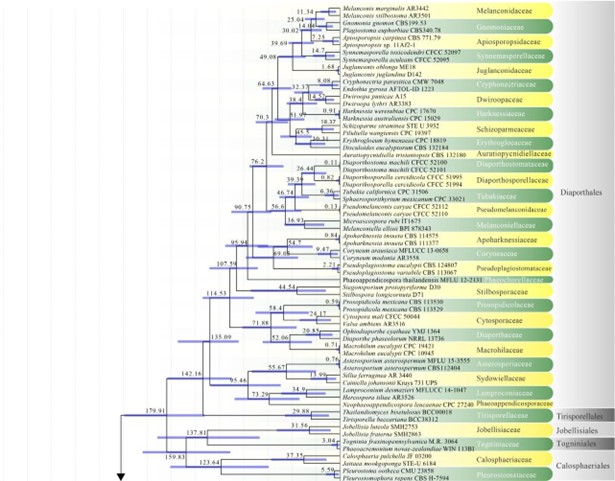
Figure 2 – The maximum clade credibility (MCC) tree, using the same dataset from Fig. 1. This analysis was performed in BEAST v1.10.2. The crown age of Sordariomycetes was set with Normal distribution, mean = 250, SD = 30, with 97.5% of CI = 308.8 MYA, and crown age of Dothideomycetes with Normal distribution mean = 360, SD = 20, with 97.5% of CI = 399 MYA. The substitution models were selected based on jModeltest2.1.1; GTR+I+G for LSU, rpb2 and SSU, and TrN+I+G for tef1 (the model TrN is not available in BEAUti 1.10.2, thus we used TN93). Lognormal distribution of rates was used during the analyses with uncorrelated relaxed clock model. The Yule process tree prior was used to model the speciation of nodes in the topology with a randomly generated starting tree. The analyses were performed for 100 million generations, with sampling parameters every 10000 generations. The effective sample sizes were checked in Tracer v.1.6 and the acceptable values are higher than 200. The first 20% representing the burn-in phase were discarded and the remaining trees were combined in LogCombiner 1.10.2., summarized data and estimated in TreeAnnotator 1.10.2. Bars correspond to the 95% highest posterior density (HPD) intervals. The scale axis shows divergence times as millions of years ago (MYA).
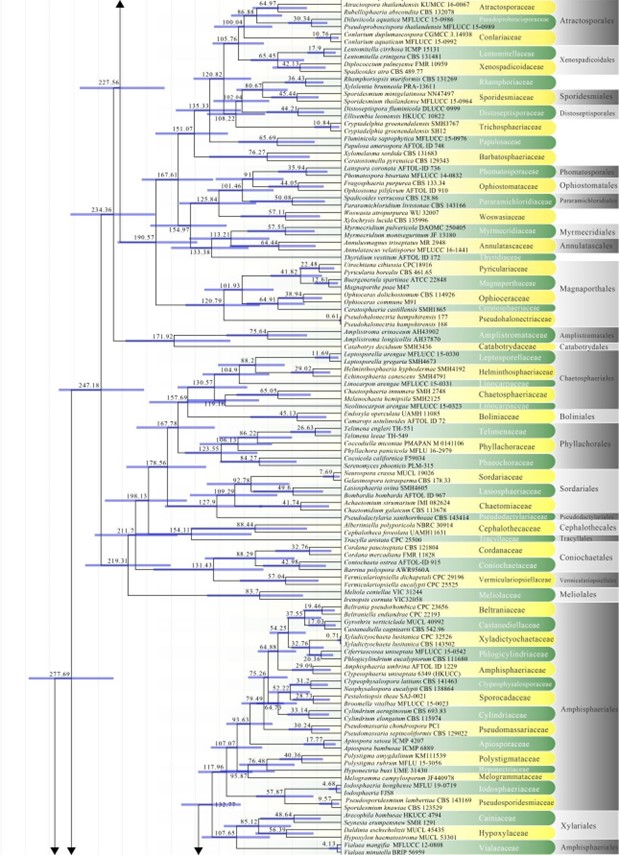
Figure 2 – Continued.
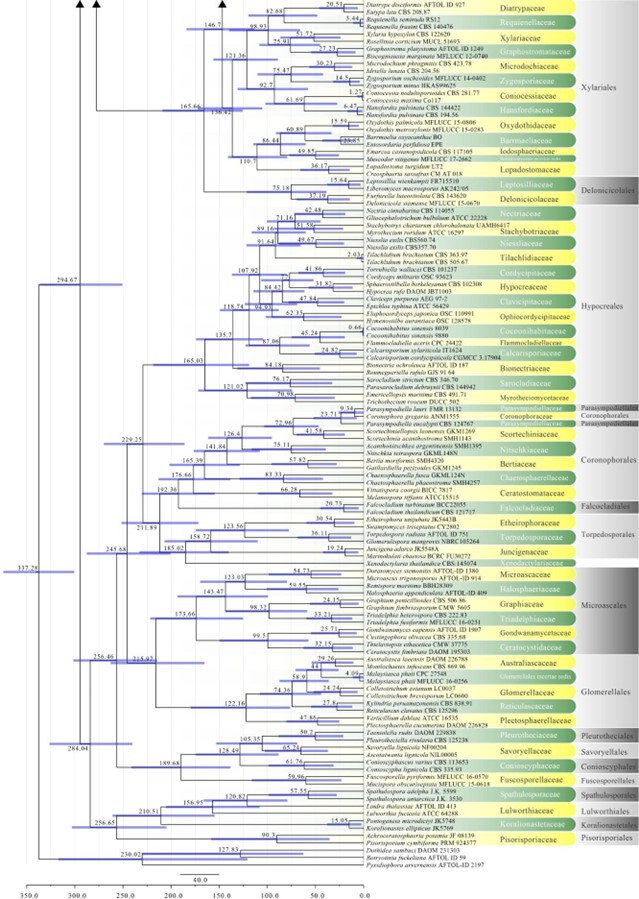
Figure 2 – Continued.

Figure 12 – Phylogram generated from maximum likelihood analysis based on combined LSU, ITS, rpb2 and tef1 sequence data of Diaporthales. One hundred and fifteen strains are included in the combined analyses which comprised 2890 characters (892 characters for LSU, 526 characters for ITS, 1062 characters for rpb2, 407 characters for tef1) after alignment. Leotia lubrica (AFTOL ID 1) is used as outgroup taxon. Single gene analyses were carried out and the phylogenies were similar in topology and clade stability. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RaxML tree with a final likelihood value of -41459.517722 is presented. Estimated base frequencies were as follows: A = 0.250714, C = 0.246950, G = 0.282507, T = 0.219829; substitution rates AC = 1.688056, AG = 2.747632, AT = 1.764840, CG = 1.180905, CT = 6.854636, GT = 1.000000; gamma distribution shape parameter a = 0.339516. Bootstrap support values for ML greater than 50% and Bayesian posterior probabilities greater than 0.90 are given near the nodes. Ex-type strains are in bold. The newly generated sequences are indicated in blue.
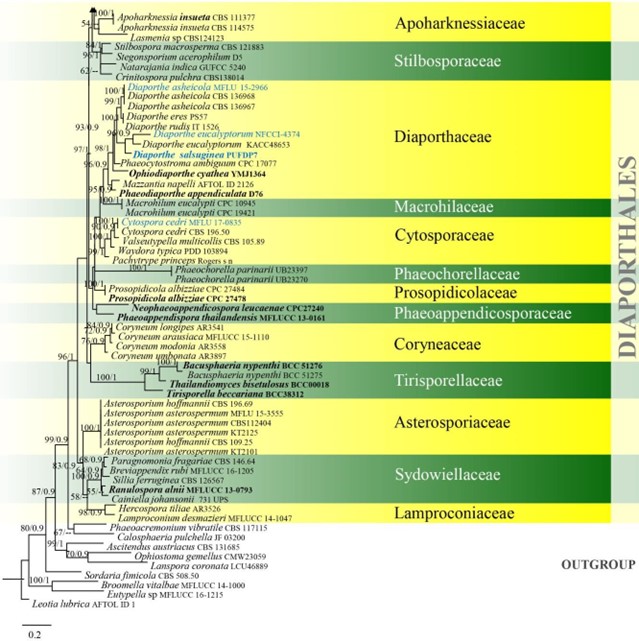
Figure 12 – Continued.
Families
