Alternaria alternata (Fr.) Keissl., Beih. bot. Zbl., Abt. 2 29: 434 (1912)
Basionym: Torula alternata Fr., Syst. mycol. (Lundae) 3(2): 500 (1832)
Pathogen or saprobe on wood and dead herbaceous stems or leaves. Sexual morph: Ascomata 170–20 × 190–240 μm (x̅ = 190 × 440 μm, n = 10), small, solitary to clustered, erumpent to (nearly) superficial at maturity, globose to ovoid, dark brown, smooth, apically papillate, ostiolate. Ostiole papilla short, blunt. Peridium 38–50 μm (x̅ = 45 μm, n = 10) wide, thin, comprising two cell types, outer layer composed of small heavily pigmented, thick-walled cells of textura angularis, inner layer composed of lightly pigmented or hyaline, thin-walled cells of textura angularis. Hamathecium of 2–2.5 μm (x̅ = 2.5 μm, n = 10) broad, long, cellular pseudoparaphyses. Asci 170–190 × 23–30 μm (x̅ = 180 × 25 μm, n = 20), (4–6–) 8-spored, bitunicate, fissitunicate, cylindrical to cylindro-clavate, straight or somewhat curved, with a short, furcate pedicel and minute ocular chamber. Ascospores 37–43 × 13–14 μm (x̅ = 37 × 13.5 μm, n = 40), ellipsoid to fusoid, slightly constricted at septa, muriform, with 3–7 transverse septa, 1–2 series of longitudinal septa through the two original central segments, end cells without septa, or with 1 longitudinal or oblique septum, or with a Y-shaped pair of septa, brown, without guttules, smooth-walled. Asexual morph: Conidiophores 40–50 × 3–6 μm solitary to clustered, simple or branched, straight or flexuous, sometimes geniculate, pale, olivaceous or golden brown, smooth. Conidia 20–63 × 9–18 μm, mostly branched, obclavate, obpyriform, ovoid or ellipsoidal, often with a short conical or cylindrical beak, pale to mid golden brown, up to 8 transverse and usually several longitudinal or oblique septa, smooth or verruculose (Ellis 1971).
Material examined: ITALY, on the dead stem, 26 March 2012, E. Camporesi IT181-1(MFLU 14-0755, ICMP) – living culture (MFLUCC 14-1184). ITALY, on the stem of Achillea sp. (Asteraceae) 15 July 2013, E. Camporesi IT 181-2 (MFLU 14-0756) – living culture (MFLUCC 14-1185). ITALY, on the stem of Portulaca sp. (Portulacaceae), 5 September 2012, E. Camporesi IT 466 (MFLU 14-0757); (KIB, PDD, isotypes).
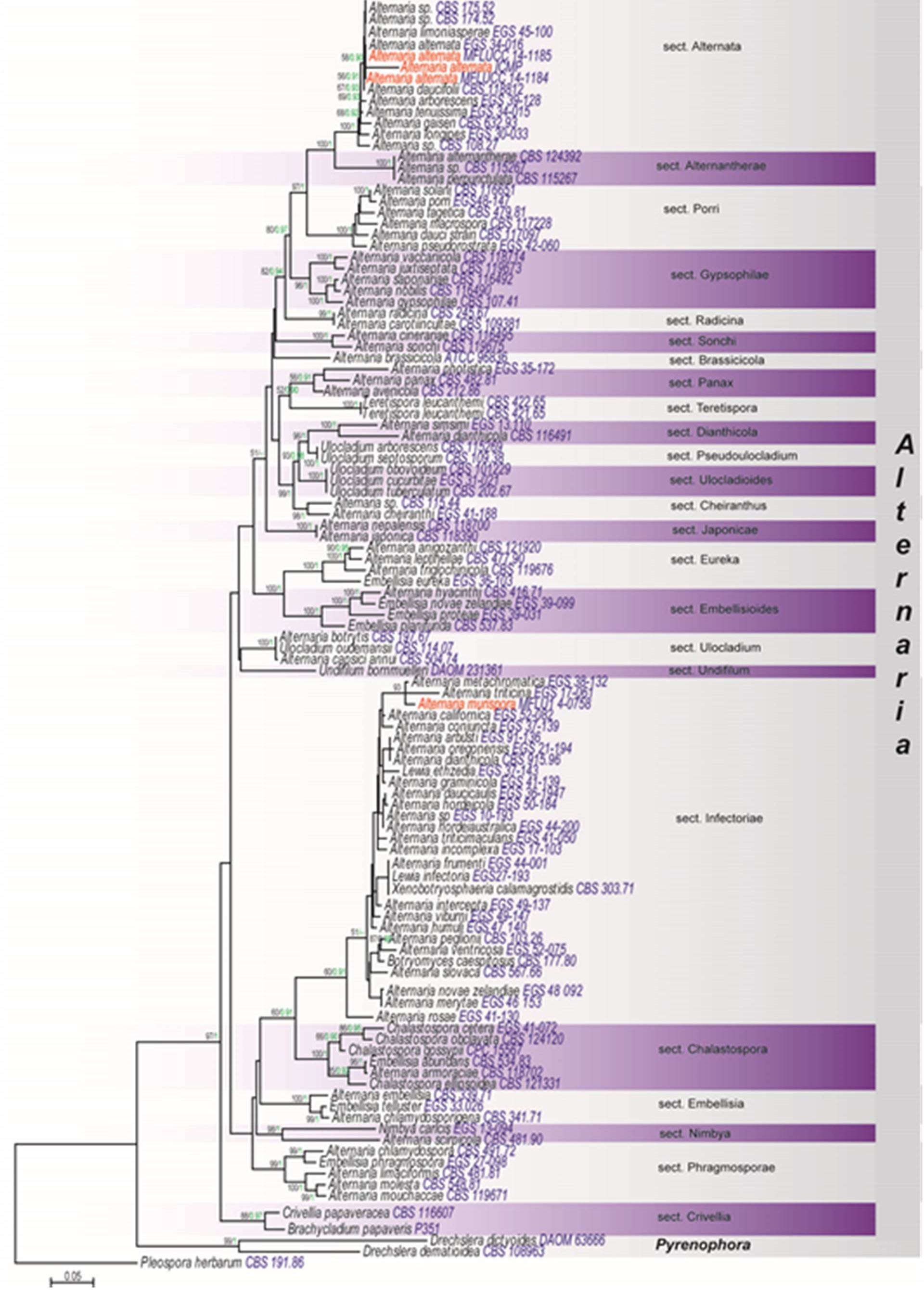
Notes: Alternaria was introduced by Nees (1816) and is a ubiquitous fungal genus that includes saprobic, endophytic and pathogenic species associated with a wide variety of substrates (Woudenberg et al. 2013). Recent studies based on DNA data as well as literature reviews revealed multiple paraphyletic genera within the Alternaria complex, and Alternaria species clades that do not always correlate to species-groups based on morphological characteristics (Woudenberg et al. 2013). Based on the combined gene analysis of GAPDH, RPB2 and TEF1, Woudenberg et al. (2013) concluded that the Alternaria clade contains 24 internal clades and six monotypic lineages, the grouping of which are recognised as Alternaria and thus synonymised Allewia, Brachycladium, Chalastospora, Chmelia, Crivellia, Embellisia, Lewia, Nimbya, Sinomyces, Teretispora, Ulocladium, Undifilum and Ybotromyces under Alternaria senso stricto Furthermore, Woudenberg et al. (2013) treated 24 internal clades in the Alternaria complex as sections. Our phylogeny based on ITS, GAPDH, LSU, RPB2 and TEF1 sequences (Fig. 1) is displayed, as these are the genes with the best resolution similar with Woudenberg et al. (2013), which is maintenance of a recent proposal for the taxonomic treatment of lineages in Alternaria.
In the present study we collected the sexual morph of Alternaria alternata from Italy in 2013 and 2014. Cultures did not produce the asexual morph but phylogenetically our collections (MFLUCC 14-1185, MFLUCC 14-1184) are 100% similar with the type strain of Alternaria alternata (CBS 916.96). Therefore here we have given the description of the sexual morph of Alternaria alternata based on our collection.
Fig 1. RAxML tree based on a combined dataset of ITS, LSU GAPDH, RPB2 and TEF1 of 114 strains representing the Alternaria-complex. Bootstrap support values for maximum likelihood greater than 50% and Bayesian posterior probabilities greater than 0.90 (green) are indicated below or above the nodes. Pleospora herbarum is the out group taxon. The original isolate numbers are noted after the species names. Newly generated strains in this study are indicated in red.