Delonicicolales R.H. Perera, Maharachch. & K.D. Hyde, Cryptog. Mycol. 38(3): 329 (2017)
MycoBank number: MB 553775; Index Fungorum number: IF 553775; Facesoffungi number: FoF 03603;
Delonicicolales was introduced by Perera et al. (2017) to accommodate Delonicicolaceae which comprises Delonicicola and Liberomyces. The divergence time estimates for the order are 181 (133–234) MYA and 95 (51–157) MYA (Perera et al. 2017). Voglmayr et al. (2019a) introduced a new genus Furfurella to Delonicicolaceae and in the same study, Liberomyces was synonymized under Leptosillia. Voglmayr et al. (2019a) also introduced Leptosilliaceae, which is typified by Leptosillia, while rejecting Delonicicolales based on phylogenetic analyses with several gene combinations (ITS-LSU and SSU-ITS-LSU-rpb1–rpb2–tef1–tub2). In our study, based on a LSU-ITS-rpb2–tub2 matrix Delonicicolales forms a highly supported clade (100% ML) comprising Delonicicolaceae and Leptosilliaceae, basal to Amphisphaeriales and Xylariales (Fig. 4). However, in another combined gene analysis of LSU-SSU-tef1–rpb2, Delonicicolales forms an internal clade in Xylariales (Fig. 1). The divergence time estimates in this study are a stem age of 165 Mya which supports the order establishment by Perera et al. (2017). Currently there are two families and three genera in this order (this paper).
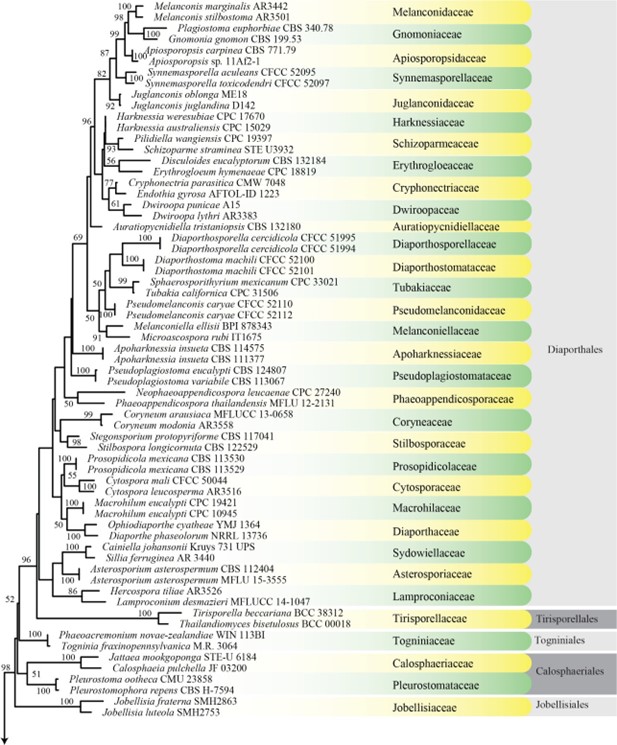
Figure 1 – Maximum likelihood (ML) majority rule combined LSU, SSU, tef1 and rpb2 consensus tree for the analyzed Sordariomycetes isolates. Families are indicated in yellow and green coloured blocks and orders are indicated in dark and light grey coloured blocks. RAxML bootstrap support values (MLB above 50 %) are given at the nodes. The scale bar represents the expected number of changes per site. The tree is rooted with Botryotinia fuckeliana (AFTOL ID-59), Dothidea sambuci (DAOM 231303), and Pyxidiophora arvernensis (AFTOL-ID 2197).
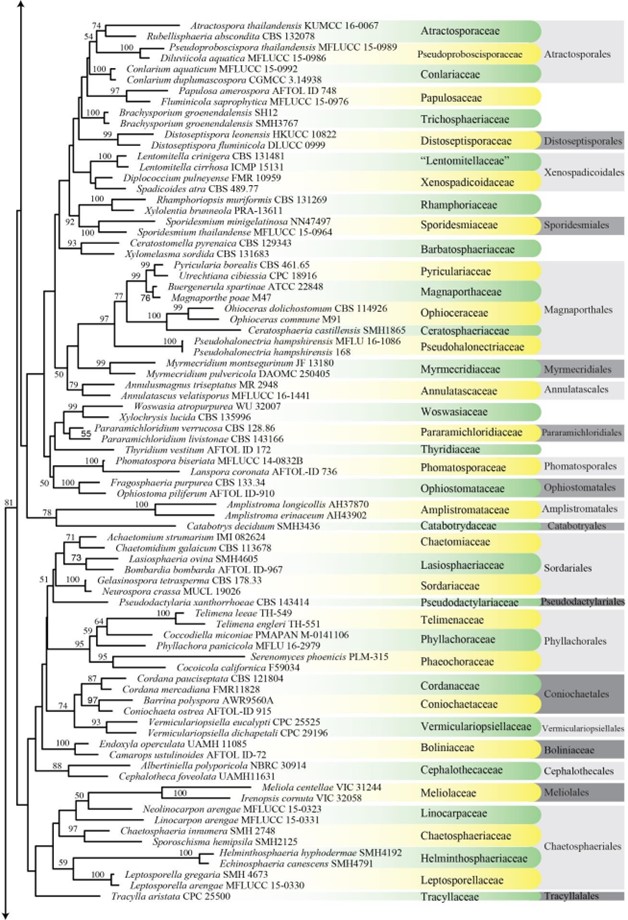
Figure 1 – Continued.
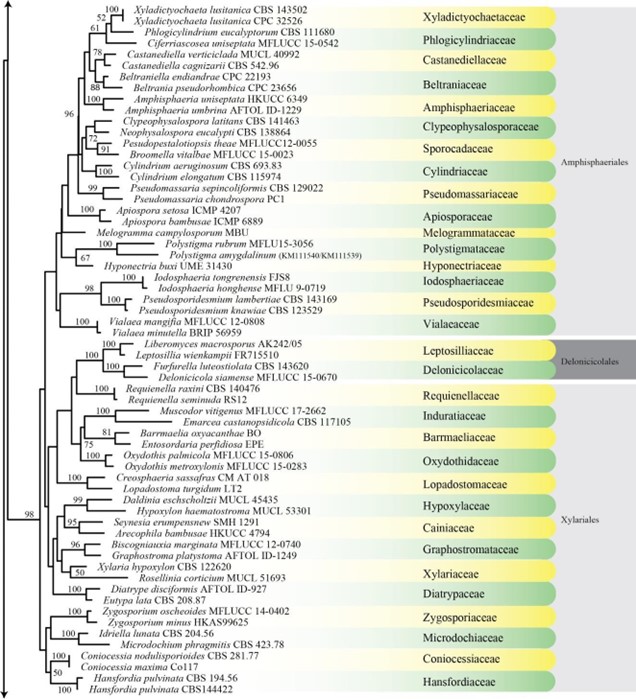
Figure 1 – Continued.
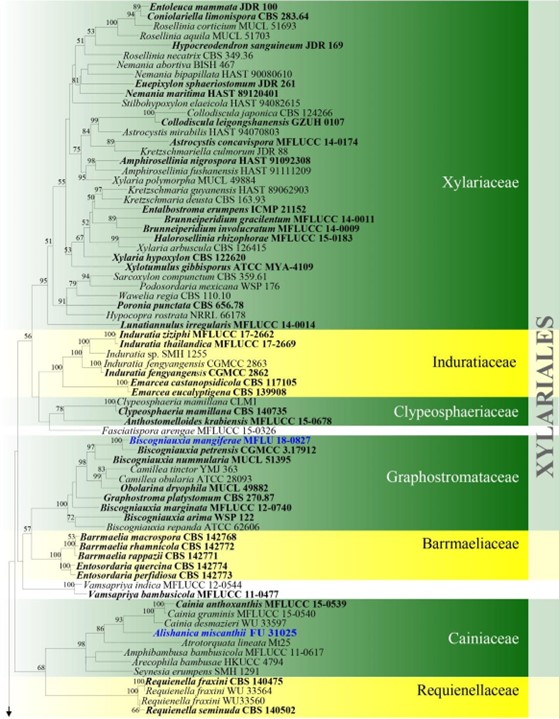
Figure 4 – Phylogram generated from maximum likelihood analysis based on combined ITS, LSU, rpb2 and tub2 sequence data for Xylariomycetidae. Two hundred and seventy-two strains are included in the combined analyses which comprised 4211 characters (1168 characters for ITS, 937 characters for LSU, 1128 characters for rpb2, 978 characters for tub2) after alignment. Achaetomium macrosporum (CBS 532.94), Chaetomium elatum (CBS 374.66) and Sordaria fimicola (CBS 723.96) are outgroup taxa. Single gene analyses were carried out and the topology of each tree had clade stability. The best RaxML tree with a final likelihood value of – 132297.706952 is presented. Estimated base frequencies were as follows: A = 0.241914, C = 0.251908, G = 0.265558, T = 0.240620; substitution rates AC = 1.281946, AG = 3.512297, AT = 1.499895, CG = 1.121065, CT = 6.472834, GT = 1.000000; gamma distribution shape parameter a = 0.678614. Bootstrap support values for ML greater than 75% are given near the nodes. Ex-type strains are in bold. The newly generated sequences are indicated in blue.
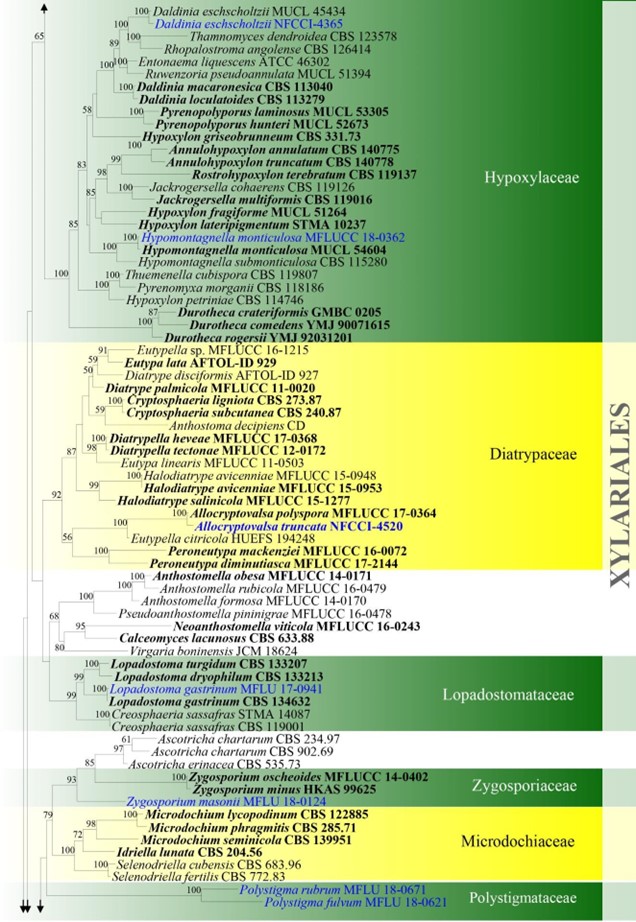
Figure – 4 Continued.
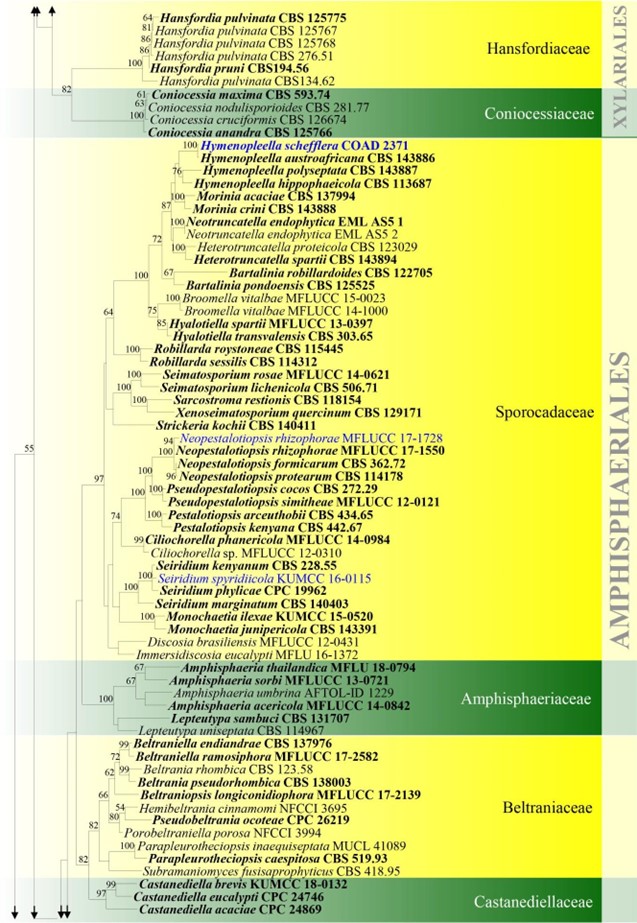
Figure – 4 Continued.

Figure – 4 Continued.
Families
