Dothideomycetes O.E. Erikss. & Winka 1997
Dothideomycetes is one of the largest and most significant classes within Ascomycota (Kirk et al. 2008; Schoch et al. 2009; Hyde et al. 2013). Thousands of species that are important either as plant pathogens in agriculture and forestry (Cortinas et al. 2006; Crous et al. 2007; Wikee et al. 2011, 2013a, b; Manamgoda et al. 2012), or medical (Siu and Lzumi 2004; da Cunha et al. 2012, 2013) or biotechnological industries (Verkley et al. 2004; Damm et al. 2008; de Wit et al. 2012; Ohm et al. 2012; Stergiopoulos et al. 2012; Hyde et al. 2014) are
included in the Dothideomycetes.
There has been considerable research interest in the molecular phylogeny of the Dothideomycetes in the last four years with studies based on single and multi-gene rDNA sequence data (Hyde et al. 2013). These phylogenetic advances have helped taxonomists to link species or generic names, enabling us to now propose or select one name for a single species, or genus.
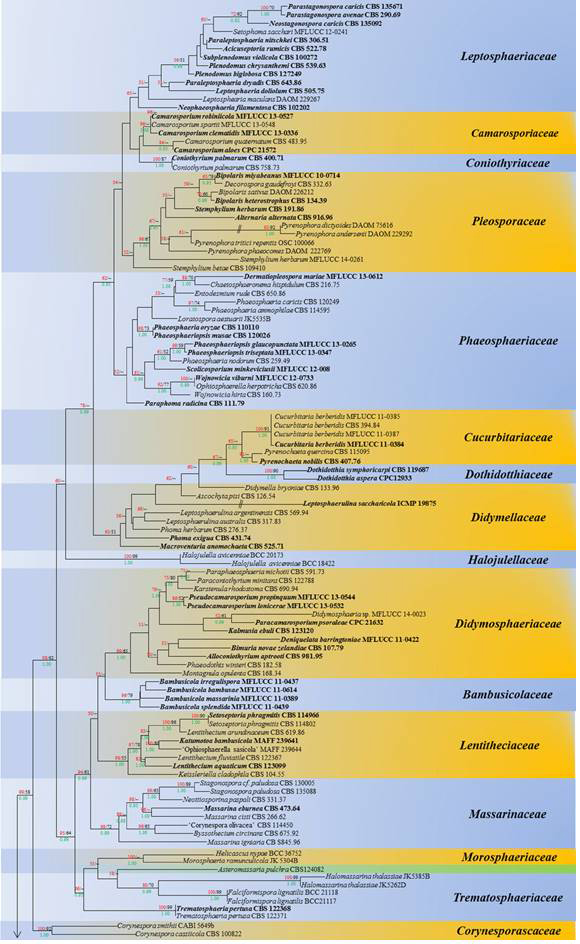
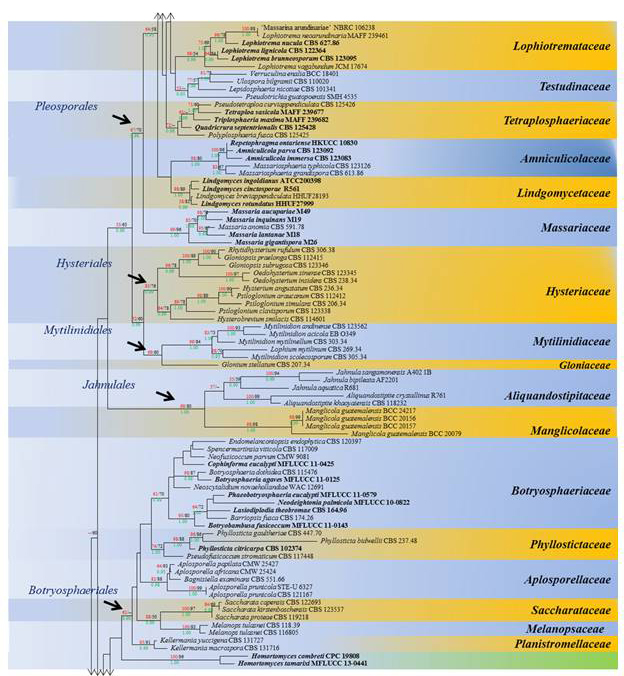
Fig. 1 The best scoring RAxML Dothideomycetes tree from 415 taxa based on a combined dataset of LSU, SSU, TEF1 and RPB2 sequences
with all lineages collapsed to family level where possible. The tree is the same as Fig. 1. Bootstrap support values for maximum likelihood (ML, red) and maximum parsimony (MP, black) greater than 50% are given above the nodes; Bayesian posterior probabilities (BYPP, green) above 0.90 are given below the nodes. The original strain numbers are given after the species names. Type and ex-type strains are emphasized in bold. The tree was rooted with Schismatomma dirinellum (S-F206034) and S. decolorans (DUKE 47570)