Luteomyces Q.V. Montoya & A. Rodrigues, gen. nov. (Fig. 5).
Index Fungorum number: MB 835150; Facesoffungi number: FoF
Etymology: “Luteomyces” based on the colour exhibited by the colonies of the type species.
Diagnosis: Similar to Escovopsis and Sympodiorosea in the way it begins to grow, i.e., dense germination and forming stolon-like mycelia. However, Luteomyces differs from these genera and other known genera in the Hypo- creaceae by its poorly differentiated holoblastic conidiog- enous cells.
Description: Monophyletic genus in the Hypocreaceae. Colonies form floccose, white, beige and yellow mycelia, stolons, beige to yellow soluble pigments. Conidiophores formed on aerial mycelia, alternated, usually arising at right angles (Fig. 5A), smooth-walled, pyramidal shape (Fig. 5B). Conidiogenous cells poorly differenciated, holoblastic, determinate, in synchronous arrangement (Fig. 5C–F) on the apices and axes of conidiophores and their branches, solitary, ampulliform to lageniform (Fig. 5B). Conidia solitary, dry, smooth or ornamented (Fig. 5G, H), yellow to light-brown. Chlamydospores abundant, hyaline, smooth (Fig. 5I).
Type species: Luteomyces trichodermoides (M. Cabello et al.) Q.V. Montoya & A. Rodrigues 2021.
Notes: – Luteomyces is phylogenetically placed in the Hypo- creaceae as a sister clade of Escovopsis (Fig. 3G). None- theless, Luteomyces grows slower and has different colony colour (mainly yellow) than Escovopsis (mainly brown), and forms conidiophores without vesicles and large num- ber of chlamydospores (rarely observed in Escovopsis).
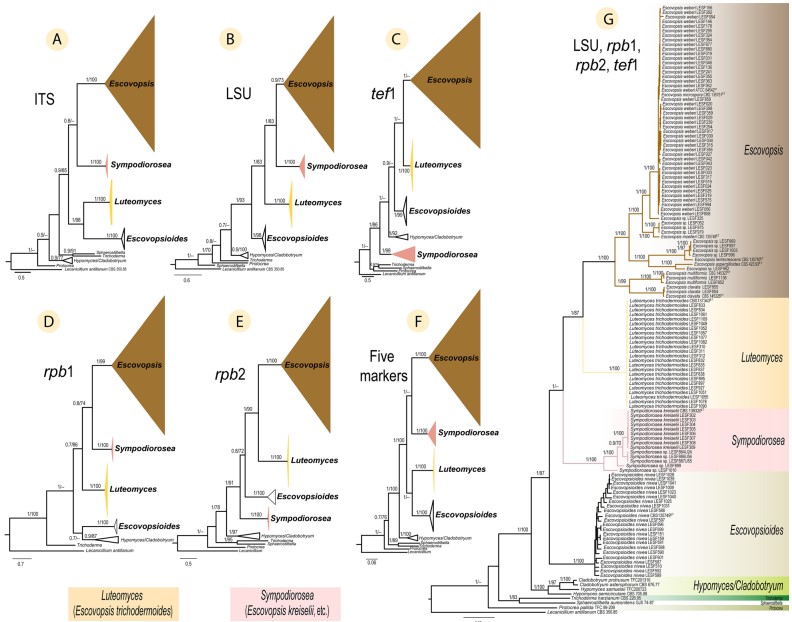
Fig. 3 Phylogenetic placement of Escovopsis, Sympodiorosea and Luteomyces within the Hypocreaceae. Phylogenies shown were inferred using Bayesian Inference (BI) and are separated for each region (molecular marker): (A) ITS, (B) LSU, (C) tef1, (D) rpb1, and (E) rpb2. The remaining two trees are based on concatenated datasets of (F) all five markers, and (G) four of the five markers (LSU, rpb1, rpb2, and tef1). Numbers on branches indicate BI posterior probabilities (PP) and Maximum Likelihood bootstrap support values (MLB), respectively. Hyphens (–) indicate MLB < 70%. Lecanicillium antillanum CBS 350.85 was used as the outgroup. ET indicates ex-type cultures. See Table S1 for all strains and their associated metadata used to infer these phylogenetic trees
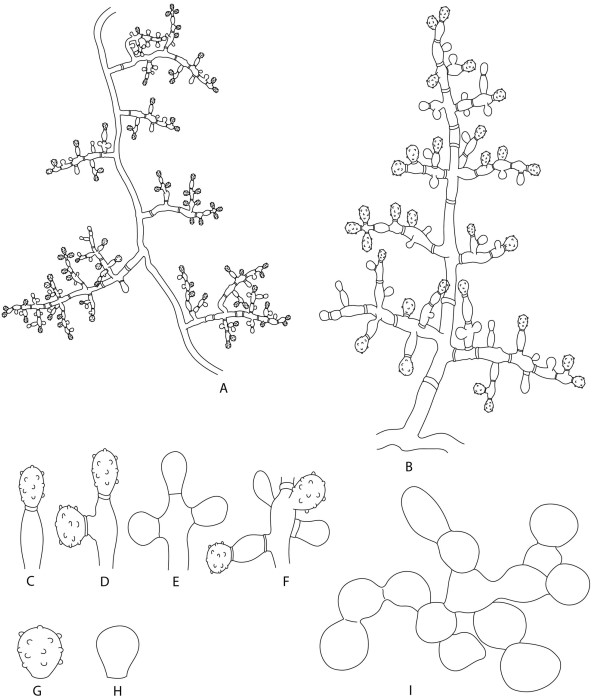
Fig. 5 Illustration of the microscopic structures of Luteomyces trichodermoides (syn. Escovopsis trichodermoides). A Pattern of disposition of conidiophores on aerial mycelium. B A conidiophore. C–F Poorly differentiated conidiogenous cells (holoblastic determinate conidiogenous cells with synchronous arrangement). G Conidium with ornamentation. H Smooth conidium. I Chlamydospores
Species
