Woswasia atropurpurea Jaklitsch, Réblová & Voglmayr, Mycologia 105(2): 479 (2013)
MycoBank number: MB 800842; Index Fungorum number: IF 800842; Facesoffungi number: FoF 11776; Fig. 26
Saprobic on dead branches of Corylus avellana L. Sexual morph: Stromata 0.8–1 mm long, 0.5–0.8 mm wide, 370–450 μm high, scattered, erumpent through the surface, surrounded by host tissues, unevenly positioned on substrate, black, multi-loculate. Ascomata perithecial, 150–200 μm diam. × 250–300 μm high (x̅ = 170 × 280 μm, n = 10), arranged in rows, immersed, obpyriform to ampulliform, dark brown to black. Ostiole raised from the centre of ascomata, lined with periphyses. Peridium 10–20 μm wide, composed of several layers, outer layer consisting of thick-walled, dark brown cells of textura angularis, inner layer of thin-walled, hyaline cells of textura angularis. Hamathecium comprising 2.5–4 μm wide, hyaline, septate, paraphyses. Asci 30–55 × 3–5 µm (x̅ = 48 × 4 µm, n = 10), 8-spored, unitunicate, cylindrical, with a slightly long pedicel, J-, apical ring. Ascospores 2.5–3.5 × 2.7–3.5 μm (x̅ = 3 × 3.2 μm, n = 20), uni-seriate, globose to subglobose, aseptate, hyaline, verruculose to smooth-walled. Asexual morph: see Jaklitsch et al. (2013).
Material examined – Italy, Province of Forlì-Cesena [FC], Teodorano – Meldola, on dead and hangingbranches of Corylus avellana L. (Fagaceae), 14 January 2019, Erio Camporesi, IT 4198, (MFLU 19-0465), ibid., 28 January 2019, IT 4198A (MFLU 19-0486).
GenBank numbers – ITS: OM616630, OM616631; LSU: OM616566, OM616567.
Notes – Morphologically, Woswasia atropurpurea is characterized by having a long neck perithecia and globose to subglobose, aseptate, and verruculose ascospores (Jaklitsch et al. 2013, this study). Morphological characters of Woswasia are remarkably similar to Amplistroma and Wallrothiella in Amplistromataceae (Jaklitsch et al. 2013). Our strains of Woswasia are morphologically similar to generic characters of Amplistroma, in having long-stipitate asci and globose to subglobose ascospores, and similar to Thalassogena, in having hyaline ascospores. However, phylogenetic analyses do not prove it (Jaklitsch et al. 2013). Ascospores were not able to germinate in PDA and MEA under different temperature conditions. According to the phylogenetic analyses, our collections (MFLU 19-0465, MFLU 19-0486) grouped with the type species W. atropurpurea (WU 32007) with 100% MLBS and 1.00 BYPP support (Fig. 27). All W. atropurpurea strains are clustered with two basal lineages within Woswasiaceae, namely Xylochrysis and Cyanoannulus (Fig. 27). Furthermore, the holotype of W. atropurpurea was found on the stromata of Diaporthe oncostoma growing on a Robinia pseudoacacia branch of Lombardia, Italy. Our strains are the first reports of W. atropurpurea on Corylus avellana from Emilia-Romagna, Italy.

Figure 26 – Woswasia atropurpurea (MFLU 19-0465). a, b. Appearance of ascomata on dead branch of Corylus avellana. c. Longitudinal sections of ascomata. d. Ostiole. e. Peridium. f. Pseudoparaphyses. g–i. Asci. j–l. Ascospores. Scale bars: a–c = 100 μm, d–f = 20 μm, g–l = 5 μm.
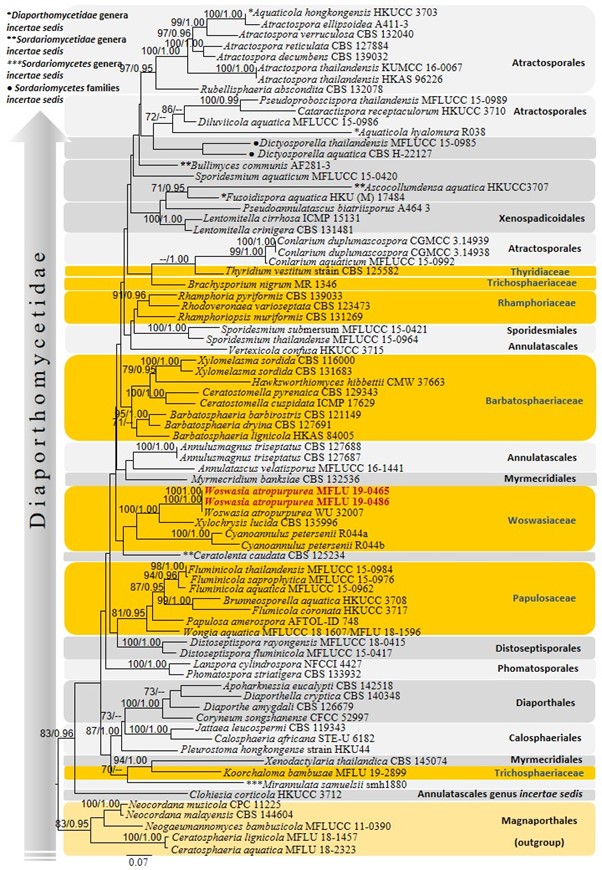
Figure 27 – Phylogram generated from maximum likelihood analysis based on combined LSU, SSU, ITS and rpb2 sequenced data. Seventy eight strains were included in the combined sequence analysis, which comprised 3286 characters with gaps (LSU = 850, SSU = 880, ITS = 528, rpb2 = 1062). Single gene analyses were also performed and topology and clade stability compared from combined gene analyses. Neocordana musicola (CPC 11225), N. malayensis (CBS 144604), Neogaeumannomyces bambusicola (MFLUCC 11-0390), Ceratosphaeria lignicola (MFLU 18-1457) and C. aquatica (MFLU 18-2323) in Magnaporthales were used as the outgroup taxa. Final ML optimization likelihood is -44234.915686. The matrix included 1836 distinct alignment patterns, with 41.80 % undetermined characters or gaps. Estimated base frequencies were obtained as follows: A = 0.250089, C = 0.241201, G = 0.286405, T = 0.222304; substitution rates AC = 1.580297, AG = 3.075761, AT = 1.536900, CG = 1.483017, CT = 7.828009, GT = 1.0; gamma distribution. Bootstrap support values for ML (first set) equal to or greater than 70% and BYPP equal to or greater than 0.95 are given above the nodes. The strains from the current study are red bold and the type strains are indicated with T.
