Vaginatispora palmae S.N. Zhang, J.K. Liu & K.D. Hyde, Fungal Diversity 96, 242 (2019)
Index Fungorum number: IF 556316; Facesoffungi number: FoF 15187; Fig. 1
Saprobic immersed on the dead trunk of Litchi chinensis. Sexual morph: Ascomata 170–270 × 220–400 µm (x̅ = 215 × 330 µm, n = 10), semi-immersed, solitary or group, globose to subglobose, dark brown to black, mastoid on top, carbonaceous, ostiolate. Ostiole central, narrow bottle pore-like opening, filled with a gelatinous substance, dark brown to black, thick-walled cells. Peridium 15–90 µm wide (x̅ = 47 µm, n = 25), think-walled, consists of 3–6 strata of slightly flattened textura angularis cells, inner layer hyaline, outer layer light brown to dark brown, thick-walled cells fused with host tissue. Hamathecium 2–4 µm wide (x̅ = 3 µm, n = 40), dense, broadly filiform, septate, thin-walled, smooth-walled, rather branched, pseudoparaphyses. Asci 60–130 × 10–20 μm (x̅ = 91 × 15 μm, n = 15), 8-spored, bitunicate, cylindrical to clavate, smooth-walled, rounded apex, with an ocular chamber and a bulbous, short pedicel. Ascospores 25–30 × 5–10 μm (x̅ = 28 × 8 μm, n = 50), overlapping biseriate, hyaline, fusiform, smooth-walled, 1-septate, slightly constricted at the middle septum, broad in the middle, tapering at both ends, apex slightly pointed or obtusely rounded, surrounded by a narrow mucilaginous sheath, with 5–10 μm long, tapering appendages, and mature ascospores with distinct oil droplets. Asexual morph: Undetermined.
Culture characteristics – Ascospores germinated on PDA within 24 hrs. Colonies on PDA reached a 20 mm diameter after two weeks at 25 °C, circular, rough surface, dense hyphae, slightly raised, extending radially outwards, surface view olivaceous brown at the middle, light olivaceous at the margins, reverse view olivaceous to dark brown at the middle, light olivaceous at the margins.
Material examined – China, Guangdong Province, Guangzhou City, Zengcheng District, on the dead trunk of Litchi chinensis (23.253964 N, 113.829763 E, 22 m), 21 September 2021, Y. H. Yang, ZGLZ030 (MHZU 22-0162), living culture ZHKUCC 22-0302.
GenBank numbers – ITS: OP735537, LSU: OP735541, SSU: OP735559, tef1-α: OQ749733, rpb2: OQ749734.
Known distribution (based on molecular data) – Thailand (Ranong: Hyde et al. 2019), China (Taiwan: Rathnayaka et al. 2021, Guangdong: this study).
Known hosts (based on molecular data) – Nypa fruticans (Hyde et al. 2019), Swietenia macrophylla (Rathnayaka et al. 2021), Litchi chinensis (this study).
Notes – In the phylogenetic analysis of the combined ITS, LSU, SSU, tef1-α and rpb2 sequence data, our new isolate (ZHKUCC 22-0302) clustered with V. palmae as a monophyletic affiliate with 99% maximum likelihood bootstrap support and 1.00 Bayesian posterior probability (Fig. 2). Except for the thicker peridium (15–90 µm vs. 15–38 µm) and longer asci (60–130 µm vs. 89–115), our collection (Fig. 1) resembles the morphology of the holotype of V. palmae (MFLUCC 18-1526) (Hyde et al. 2019). Based on morphological and phylogenetic similarities, we described herein our new isolate (ZHKUCC 22-0302) as a new host record of V. palmae, inhabiting the dead trunks of Litchi chinensis in China.
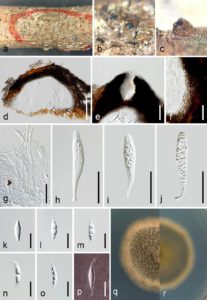
Fig. 1– Vaginatispora palmae (MHZU 22-0162, a new host record). a–c Ascomata on the host substrate. d Vertical section of an ascoma. e Prolonged neck. f Peridium. g Pseudoparaphyses. h–j Asci. k–o Ascospores. p Ascospore stained with Indian Ink. q, r Upper (q) and reverse view (r) of the colony on PDA. Scale bars: d = 100 µm, e, f, h–j = 50 µm, k–p = 20 µm, g = 30 µm.
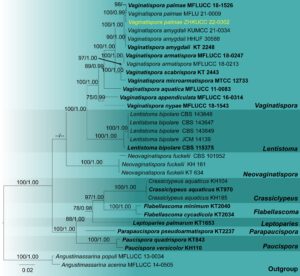
Fig. 2 – Phylogram generated from maximum likelihood analysis based on the combined ITS, LSU, SSU, tef1–α and rpb2 sequence data of the genus Vaginatispora. Thirty-two isolates are included in the combined analyses which comprised 4575 characters (806 characters for ITS, 853 characters for LSU, 986 characters for SSU, 901 characters for tef1–α and 1029 characters for rpb2). Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RaxML tree with a final likelihood value of -16632.948483 is presented. The matrix had 1094 distinct alignment patterns, with 22.27% of undetermined characters or gaps. The GTR+G evolutionary model was applied to all the genes. Bootstrap support values for ML greater than or equal to 75% and Bayesian posterior probabilities greater than or equal to 0.95 are given near the nodes, respectively. The tree was rooted with Angustimassarina populi (MFLUCC 13-0034) and Angustimassarina acerina (MFLUCC 14-0505). Ex-type strains are in bold. The newly generated sequences are indicated in yellow
