Pseudopithomyces chartarum (Berk. & M. A. Curtis) Jun F. Li, Ariyaw. & K.D. Hyde, in Ariyawansa et al., Fungal Diversity 75, 27–274 (2015)
Index Fungorum number: IF 551393; Facesoffungi number: FoF 00938; Fig. 1
Saprobic on the leaves of Musa sp. Sexual morph: Undetermined. Asexual morph: Hyphomycetous. Colonies scattered, powdery, dark brown to black, effuse. Vegetative hyphae hyaline to pale brown, septate, branched. Conidiophores 7–10 µm × 4–5 µm (x̅ = 9 × 4.5 µm, n = 20), micronematous, mononematous, hyaline, unbranched, septate or aseptate, thin-walled, with globular base and a truncated apex. Conidiogenous cells 5–8 µm × 3–5 µm (x̅ = 7.8 × 4.6 µm, n = 20), terminal, hyaline, globose or subglobose, integrated, hyaline. Conidia 18–25 µm × 10–15 µm (x̅ = 19.5 × 12 µm, n = 20), subglobose, initially light brown, becoming brown to dark brown at maturity, 3–4 transverse septa, 1–3 longitudinal septa, darken and slightly constricted at the septa, verruculose, thick-walled.
Culture characteristics – Colonies on PDA, reaching 5 cm diam. at 14 days at room temperature (25–30 ℃), superficial, cottony, pinkish white, radially striate with a regular edge.
Material examined – Thailand, Chiang Rai Province, Rattana Dormitory, dead leaf of Musa sp., 20 May 2020, Binu C. Samarakoon, BNS246 (MFLU 19-0408), living cultures MFLUCC 17-2567.
GenBank numbers – ITS: OR186205, LSU: OR186208, tef 1-α: OR195686.
Known distribution (based on molecular data) – worldwide (Lumyong et al. 2003, Ariyawansa et al. 2015)
Known hosts (based on molecular data) – dead leaf of Musa sp. (Lumyong et al. 2003, Ariyawansa et al. 2015, this study)
Notes – Our isolate was identified as Pseudopithomyces chartarum based on the morphology and multi-gene phylogeny of combined SSU, LSU, ITS, and tef 1-α. In the multi-gene phylogeny, our isolate MFLUCC 17-2567 clustered with Pseudopithomyces chartarum with 99% maximum likelihood bootstrap support and 1.00 Bayesian posterior probability (Fig. 2). However, our species shows similar morphological characters (Fig. 1) to the type species except for a few differences, such as the shapes and sizes of conidiophores and conidia (Ariyawansa et al. 2015). These changes may be due to environmental influences. Here, we provide a saprobic collection of P. chartarum from dead Musa sp. in Thailand.

Fig. 1 Pseudopithomyces chartarum (MFLU 19-0408, a new saprobic collection). a Colonies on dead banana leaf. b–c Conidiophores bearing conidia. d–h Conidia. i Colony on the PDA. Scale bars: a = 0.1 mm, b–g = 15 µm, h = 18 µm.
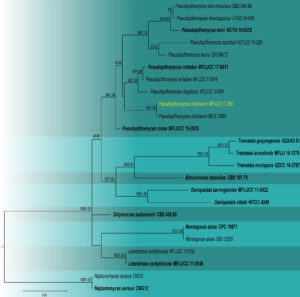
Fig. 2 Phylogram generated from maximum likelihood analysis based on combined SSU, LSU, ITS, tef1-α sequence data for the genus Pseudopithomyces and related taxa. Twenty-four strains are included in the combined analyses which comprised 2972 characters (916 characters for SSU, 730 characters for LSU, 400 characters for ITS and 914 characters for tef1-α) after alignment. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RaxML tree with a final likelihood value of -14172.708958 is presented. The matrix had 450 distinct alignment patterns, with 34.59% of undetermined characters or gaps. The evolutionary model SYM+I+G applied to LSU sequence data, while GTR+I+G applied to ITS, SSU and tef1-α gene regions. Bootstrap support values for ML equal to or greater than 80% and Bayesian posterior probabilities equal to or greater than 0.95 are given near nodes, respectively. The tree is rooted with Neptunomyces aureus (CMG 12, CMG 13). Ex-type strains are in bold. The newly generated sequences are indicated in yellow.
