Nigrograna magnoliae Wanas., in Wanasinghe, Wijayawardene, Xu, Cheewangkoon & Mortimer, PLoS ONE 15(7, e0235855), 10 (2020)
Index Fungorum number: IF 557331; Facesoffungi number: FoF 09092; Fig. 1
Saprobic on dead branches of Michelia alba, Rosa sp. and fungal fruiting bodies of Shearia sp. Sexual morph: Ascomata 220–365 × 190–285 μm (x̅ = 289 × 249 μm, n = 8), perithecioid, singly gregarious, immersed in bark or fungal fruiting bodies, sometimes erumpent, subglobose or ellipsoidal, black, with an ostiole, cylindrical neck. Ostiole mostly central, brittle. Peridium 15–22 μm (x̅ = 18.7 μm, n = 10) wide, 3–4-layered, composed of textura angularis cells, outer layer, dark brown, thick-walled, large cells, often surrounded by an olivaceous to dark brown subiculum, middle and inner layers, pale brown, thin-walled small cells. Hamathecium composed of 1.5–2.7 μm (x̅ = 2.3 μm, n = 25) wide, numerous, tapering distally pseudoparaphyses, rarely trabeculate pseudoparaphyses. Asci 70–86 ×8.5–10.5 μm (x̅ = 78 × 9.5 μm, n = 20), bitunicate, fissitunicate, 8-spored, clavate to cylindric-clavate, short pedicellate, apically rounded, sometimes with an ocular chamber. Ascospores 13.5–18.5 × 4.4–6.5 μm (x̅ = 15.8 × 5.6 μm, n = 25), biseriate, fusoid to clavate, yellowish brown to dark brown, 3-septate, apical cell often acute, second cell usually slightly widened, constricted at the medium, only slightly at other septa, straight or curved, terminal cells often slightly longer than mid cells. Asexual morph: Undetermined.
Materials examined – China, Guizhou, Guiyang, Guizhou Academy of Agricultural Sciences (GZAAS) premises, on dead branch of Michelia alba, and fruiting bodies of Shearia sp., 7 October 2019, MC Samarakoon SAMC266 (HKAS 107035); China, Sichuan, Chengdu, UESTC premises, on dead branch of Rosa sp., 30 September 2019, MC Samarakoon SAMC259 (HKAS 107027).
GenBank accession numbers – ITS: OM293747, LSU: OM293741, SSU: OM293752, rpb2: OM305055, β-tubulin: OM305065, tef1-ɑ: OM305059 (HKAS 107035); ITS: OM293746, LSU: OM293740, SSU: OM293751, β-tubulin: OM305064, tef1-ɑ: OM305058 (HKAS 107027).
Known distribution (based on molecular data) – China (Wanasinghe et al. 2020, Zhang et al. 2020, this study), Thailand (Zhang et al. 2020).
Known hosts (based on molecular data) – Magnolia denudata (Wanasinghe et al. 2020), Michelia alba, Rosa sp. and fruiting bodies of Shearia sp. (this study), and several unidentified hosts (Zhang et al. 2020).
Notes – Wanasinghe et al. (2020) discovered Nigrograna magnoliae with sexual and asexual morphs on living Magnolia denudata branches. Zhang et al. (2020) described two collections of N. magnoliae on decaying twigs of an unidentified host and submerged wood from China and Thailand, respectively. The size of the asci and ascospores of the type strain (MFLU 20–0092) overlaps with those of our new collections (asci: 60–100 × 8–12 μm vs. 70–86 × 8.5–10.5 μm and ascospores: 12–16 × 5–6.5 μm vs. 13.5–18.5 μm × 4.4–6.5 μm). Our new collections are morphologically and phylogenetically similar to N. magnoliae and are introduced as new records. Interestingly, we observed some ascomata of N. magnoliae immersed in conidiomata and ascomata of Shearia sp. (Fig. 20).

Fig. 1 – Nigrograna magnoliae (HKAS 107035 and HKAS 107027, new host records). a, c, d Ascomata on Michelia alba dead branches. b, e, f Ascomata on Rosa sp. dead branches (white arrows show ascomata). g Cross section of an ascoma. h Section of the peridium. i Pseudoparaphyses. j–m Asci. n–s Ascospores. Scale bars: a, b = 1 cm, c = 1000 μm, e = 500 μm, d, f = 200 μm, g = 100 μm, h, j–m = 20 μm, n–s = 10 μm, i = 5 μm.
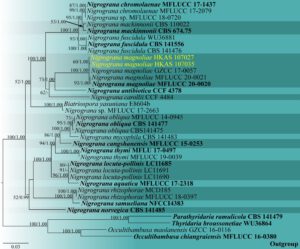
Fig. 2 – Phylogram generated from maximum likelihood analysis based on the combined LSU, SSU, tef1-α, and rpb2 sequence data of the genus Nigrograna. Thirty-six isolates are included in the combined analyses which comprised 3799 characters (833 characters for LSU, 1023 characters for SSU, 933 characters for tef1-α and 1010 characters for rpb2). Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RaxML tree with a final likelihood value of -14937.288660 is presented. The matrix had 951 distinct alignment patterns, with 25.69% of undetermined characters or gaps. The GTR+G+I evolutionary model was applied for all the genes. Bootstrap support values for ML equal to or greater than 50% and Bayesian posterior probabilities equal to or greater than 0.95 are given near the nodes, respectively. The tree was rooted with Occultibambusa chiangraiensis (MFLU 16-0380), O. maolanensis (GZCC 16-0116), Parathyridaria ramulicola (CBS 141479) and Thyridaria broussonetiae (WU36864). Ex-type strains are in bold. The newly generated sequences are indicated in yellow
