Scortechiniaceae Huhndorf, A.N. Mill. & F.A. Fernández, Mycol Res 108(12): 1387 (2004)
MycoBank number: MB 82146; Index Fungorum number: IF 82146; Facesoffungi number: FoF 01123; 33 species.
Saprobic on woody substrates in terrestrial habitats. Sexual morph: Ascomata perithecial, scattered or gregarious, superficial, semi-immersed or immersed in host, sitting on or in a subiculum or subiculum absent, globose, obpyriform or ovoid, dark brown to black, coriaceous to membranous, turbinate or tuberculate or smooth, with or without brown branched setae, collabent or not collapsing. Subiculum thin or thick, brown to dark brown, septate, crisp when dry, branched or unbranched hyphae, with spines or smooth. Peridium thick, Munk pores present or absent, outer layer composed of black to brown cells of textura angularis, thick-walled; inner layer composed of hyaline cells of textura prismatica, thin-walled. Hamathecium Quellkörper present, conical, wide at the base, paraphyses absent or indistinct, filiform, hyaline. Asci 8- to multi-spored, unitunicate, cylindrical to clavate, long or short pedicellate, apical ring J-, indistinct or absent, evanescent. Ascospores 2-seriate to irregularly arranged, hyaline or light brown, allantoid, ellipsoid to oval, straight to slightly curved, aseptate, with or without appendages. Asexual morph: Conidiophores erect, solitary, straight to flexuous. Conidiogenous cells hyaline, smooth, subcylindrical. Conidia aseptate, hyaline, smooth, ellipsoid to ovoid, granular (adapted from Huhndorf et al. 2004a).
Type genus – Scortechinia Sacc.
Notes – Scortechiniaceae was introduced by Huhndorf et al. (2004a) based on superficial, black, turbinate ascomata with a quellkörper and clavate asci with hyaline ascospores, as well as on LSU sequence analyses. The Quellkörper is believed to play a role to rupture the ascoma and probably is involved in ascospore discharge (Mugambi & Huhndorf 2010). This structure was accepted as a main character to define this family when other morphs were not significant (Huhndorf et al. 2004a, Mugambi & Huhndorf 2010, Carneiro de Almeida et al. 2016). Huhndorf et al. (2004a) accepted Euacanthe, Neofracchiae and Scortechinia as the members in the family based on LSU sequence data. Mugambi & Huhndorf (2010) added Biciliospora, Coronophorella, Cryptosphaerella, Scortechiniella, Scortechiniellopsis, and Tympanopsis in Scortechiniaceae based on multi-gene phylogenetic analyses. Crous et al. (2013) introduced the monotypic genus, Pseudocatenomycopsis, which closely related to Neofracchiaea, Cryptosphaerella and Scortechiniellopsis based on LSU sequence analyses. Members of Scortechiniaceae constitute a strongly supported monophyletic group in Fig. 11. In this entry we illustrate Neofracchiaea callista. Most members of this family have molecular support for their inclusion, but several of the genera are monotypic.
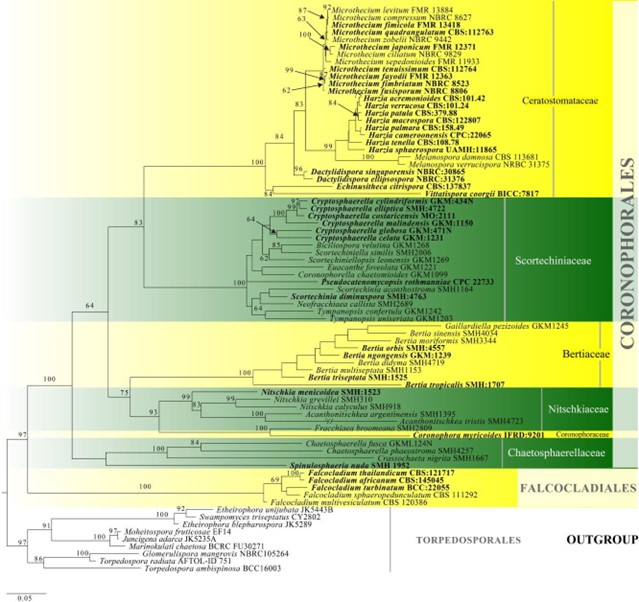
Figure 11 – Phylogram generated from maximum likelihood analysis based on combined LSU, tef1 and ITS sequence data for Coronophorales. Related sequences are referred to Hongsanan et al. (2017). Seventy-nine strains are included in the combined analyses which comprised 2211 characters (1044 characters for LSU, 627 characters for tef1, 540 characters for ITS) after alignment. Members of Torpedosporales are used as outgroup taxa. Single gene analyses were carried out and the phylogenies were similar in topology and clade stability. The best RaxML tree with a final likelihood value of -23354.525618 is presented. Estimated base frequencies were as follows: A = 0.228019, C = 0.288494, G = 0.281955, T = 0.201533; substitution rates AC = 1.194725, AG = 2.787601, AT = 1.703067, CG = 0.955075, CT = 3.781057, GT = 1.000000; gamma distribution shape parameter a = 0.736138. Bootstrap support values for ML greater than 50% are given near the nodes. Ex-type strains are in bold. The newly generated sequences are indicated in blue.
Genera
