Neocryptosphaerella S.K. Huang & K.D. Hyde, gen. nov.
Etymology: Name refers to genus being similar to Cryptosphaerella.
Index Fungorum number: IF558354; Facesoffungi number: FoF 10165
Saprobic on wood.
Sexual morph: Ascomata solitary or scattered, immersed to erumpent, black, turbinate, the apex collapsing when dry, tuberculate, sitting in a subiculum, lacking ostioles, with a central, conical Quellkörper. Peridium composed of brown to hyaline cells of textura angularis to textura prismatica, Munk pores present. Paraphyses absent. Asci polysporous, unitunicate, clavate, with long pedicel, apex rounded, without apical ring, evanescent. Ascospores overlapping, hyaline, allantoid to cylindrical, slightly curved, aseptate, smooth-walled, guttulate.
Asexual morph: Undetermined (adapted from Mugambi & Huhndorf 2010).
Notes: – We establish Neocryptosphaerella based on Cryptosphaerella celata (type) and C. globosa. Mugambi & Huhndorf (2010) introduced six ‘Cryptosphaerella’ species having ascomata lacking ostioles and a Quellkörper. The characteristics of the type of Cryptosphaerella are ostiolate ascomata lacking a Quellkörper (Saccardo 1882a) (see notes for Cryptosphaerella). Therefore, Cryptosphaerella should not belong to Scortechiniaceae and these six species do not belong to Cryptosphaerella based on morphology. Phylogenetically, C. celata and C. globosa form an independent clade and the remaining four species form another clade in Scortechiniaceae (Mugambi & Huhndorf 2010). The phylogenetic position of the type of Cryptosphaerella is undetermined. We therefore introduce Neocryptosphaerella for C. celata and C. globosa. This genus is sister to Biciliospora, Coronophorella, Pseudocryptosphaerella, Scortechiniella and Scortechiniellopsis in Scortechiniaceae, and has ascomata lacking ostioles, Quellkörper and polysporous asci with allantoid to cylindrical ascospores (Mugambi & Huhndorf 2010, this study, 0.98BY, Fig. 1).
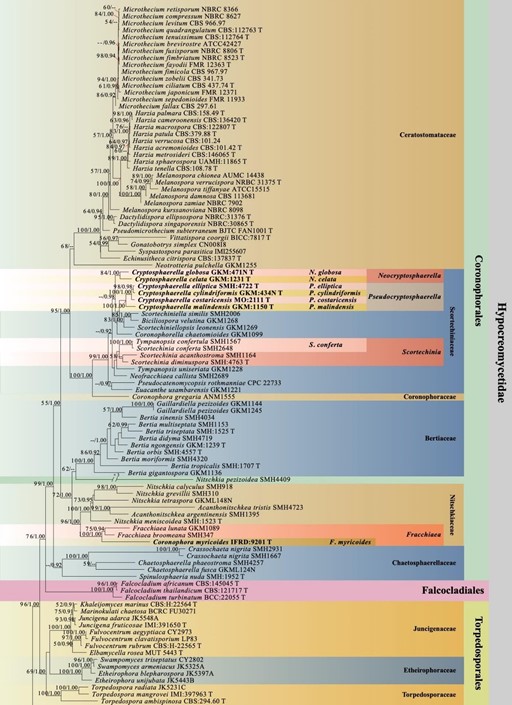
Figure 1 – Phylogram generated from maximum likelihood analysis based on combined LSU, SSU, TEF, RPB2, TUB and ITS sequence data with the confidence values of bootstrap (BS) proportions from the Maximum Likelihood (ML) analysis (ML-BS > 50%, before the backslash) and the posterior probabilities (PP) from the Bayesian (BY) analysis (BY-PP > 0.90, after the backslash) above corresponding nodes. The ‘–’ indicates a lack of statistical support (< 50% for ML-BS and < 0.90 for BY-PP). One hundred and ninety-eight strains are included in the combined analyses which comprised 4581 characters (842 characters for LSU, 1025 characters for SSU, 950 characters for TEF, 876 characters for RPB2, 391 characters for TUB, 497 characters for ITS) after alignment. Strains of Xylariomycetidae are used as outgroup taxa. The best score in IQ-TREE explores with a final likelihood value of -90984.2293 is presented. The model of each partitioned gene is LSU: GTR+I+G; SSU: GTR+G; TEF: GTR+I+G; RPB2: GTR+I+G; TUB: GTR+I+G; ITS: GTR+G. Sequences generated indicated in bold. Strain numbers are noted after the species names and ex-type strains marked with ‘T’ after the culture number. Alignment is available at TreeBASE (URL: http://purl.org/phylo/treebase/phylows/study/TB2:S28271).
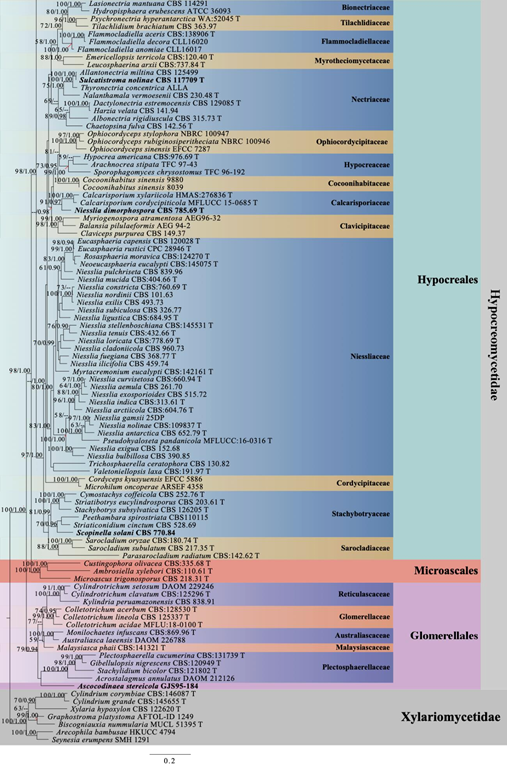
Figure 1 – Continued.
Species
