Sarcoscyphaceae Le Gal ex Eckblad, Nytt Mag. Bot. 15(1-2): 103 (1968)
Index Fungorum number: IF 81256
This family consists of mostly saprobic species. There are no documented hypogeous taxa (Pfister 2015). The family is characterized by vivid apothecia, pigmented paraphyses, suboperculate asci, and smooth or ornamented ascospores (Ekanayaka et al. 2018). These species abundantly occur in tropical areas (Pfister 2015). Fourteen genera are accepted by Ekanayaka et al. (2018), while the updated outline of fungi listed 12 genera in this family (Wijayawardene et al. 2020). An updated phylogeny is provided in Fig. 30.
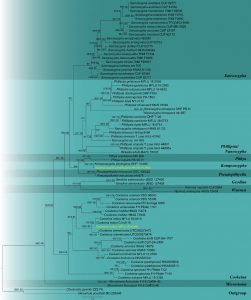
Phylogram generated from maximum likelihood analysis based on combined ITS and LSU sequence data. Seventy-five strains are included in the combined analyses which comprised 1479 characters (604 characters for ITS, 875 characters for LSU) after alignment. The GTR+I+R was applied as the evolutionary model for all the gene regions. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RaxML tree with a final likelihood value of -14833.676115 is presented. Estimated base frequencies were as follows: A = 0.230206, C = 0.253698, G = 0.280642, T = 0.235454; substitution rates AC = 1.200880, AG = 2.035026, AT = 1.401259, CG = 1.177486, CT = 4.247817, GT = 1.000000; gamma distribution shape parameter α = 0.330664. Bootstrap support values for ML greater than 75% and Bayesian posterior probabilities greater than 0.95 are given near nodes respectively. The tree is rooted with Chorioactis geaster (ZZ2 FH) and Neournula pouchetii (MO 205345). The newly generated sequences are indicated in yellow
Genera
Aurophora
Geodina
Microstoma
Nanoscypha
Phillipsia
Pithya
Pseudopithyella
Sarcoscypha
Thindia
Wynnea
