Pseudoconlarium N.G. Liu, K.D. Hyde & J.K. Liu, gen. nov.
Index Fungorum number: IF557094; Facesoffungi number: FoF 06705
Etymology: Referring to the morphologically similar genus Conlarium.
Saprobic on decaying wood.
Sexual morph: Unde- termined.
Asexual morph: Colonies on natural substrate effuse, scattered, punctiform. Mycelium partly immersed, partly superficial. Conidiophores micronematous or semi- macronematous, mononematous, septate, branched, flexu- ous, subhyaline to pale brown. Conidiogenous cells mono- blastic, integrated, terminal, subhyaline to pale brown, determinate, doliiform. Conidia solitary, brown, irregularly globose to subglobose, or trapeziform, muriform, constricted at the septa, smooth-walled.
Type species: Pseudoconlarium punctiforme N.G. Liu,K.D. Hyde & J.K. Liu
Notes: – Pseudoconlarium resembles Conlarium in having punctiform colonies, micronematous or semi-macronema- tous conidiophores, and subglobose and muriform conidia. However, in our phylogenetic analyses based on combined LSU, SSU, ITS, TEF1-α sequence data (Fig. 120), Pseudo- conlarium did not fit in with any orders or families in the subclass Diaporthomycetidae and its phylogenetic placement is unstable. Thus, we introduce a new genus and treat it as Diaporthomycetidae genus incertae sedis in this study.
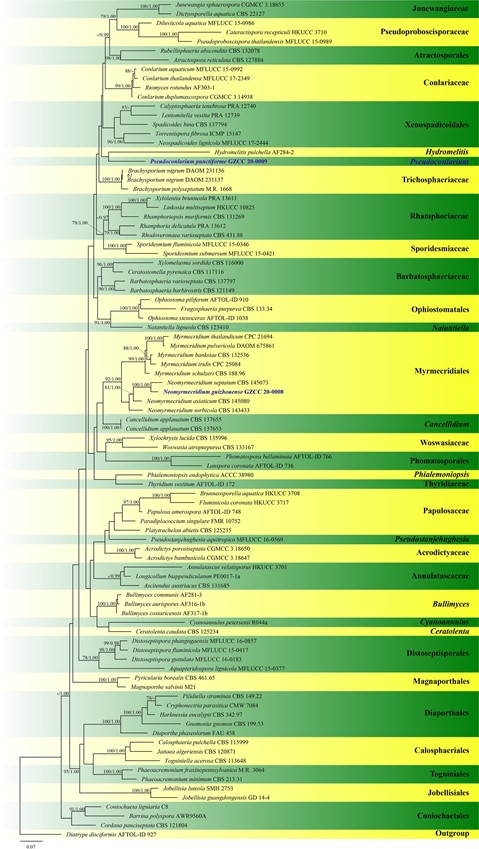
Fig. 120 Phylogram generated from maximum likelihood anal- ysis based on combined LSU, SSU, ITS and TEF1-α sequence dataset representing Diaportho- mycetidae (Sordariomycetes). Ninety-one strains are included in the combined analyses which comprise 4690 characters (1203 characters for LSU, 1646 char- acters for SSU, 712 characters for ITS, 1129 characters for TEF1-α) after alignment. Dia- trype disciformis (AFTOL-ID 927) in Diatrypaceae (Xylari- ales) is used as the outgroup taxon. Single gene analyses were also performed to compare the topology and clade stability with combined gene analyses. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RAxML tree with a final likeli- hood values of − 54946.205991 is presented. The matrix
had 2477 distinct alignment patterns, with 49.48% unde- termined characters or gaps. Estimated base frequencies were as follows: A = 0.248881, C = 0.237137, G = 0.282063, T = 0.231918; substitution rates AC = 1.401634, AG = 2.637969, AT = 1.281525, CG = 1.264278, CT = 5.804548, GT = 1.000000;
gamma distribution shape parameter α = 0.240704. Bootstrap values for maximum likelihood (ML) equal to or greater than 75% and clade credibility values greater than 0.95 from Bayesian-inference analysis labeled on the nodes. The new isolates are indicated in bold and blue
Species
