Phyllosticta chiangmaiensis Gomdola D & Hyde K.D. sp. nov. Fig. 1
MycoBank number: MB; Index Fungorum number: IF; Facesoffungi number: FoF 12965;
Etymology – The specific epithet refers to the location where the specimen was collected.
Holotype – MFLU XXX FoF 12965
Saprobic on Musa sp. Sexual morph Not observed. Asexual morph Coelomycetous. Conidiomata 50–160 × 50–160 µm (x̄=90 × 95 μm, n=20), solitary, uniloculate, globose to sub-globose, scattered or gregarious, semi-immersed, conspicuous on host surface, black. Pycnidial walls 2.75–14.5 μm wide (x̄=7.71 μm, n=30), comprising 1–2 layers of thin-walled textura angularis cells, outer layer dark brown to black, inner layer pale brown. Ostiole not observed. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 6.0–14.0 × 4.5–9.0 μm (x̄=9.82 × 6.39 µm, n=25), sub-globose or ellipsoidal or ovoid, guttulate, aseptate, hyaline, smooth. Conidia 13.0–18.0 × 8.5–12.5 µm (x̄=15.5 × 9.95 μm, n=60), solitary, ellipsoidal to obovoid, guttulate, verruculose when immature with a single large central guttule when mature, aseptate, hyaline, smooth-walled, tapering towards a narrowly truncate base and broader apex, surrounded by a mucilaginous sheath, thicker on both sides, 1.20–4.70 µm thick (x̄=2.61 μm, n=60), thinner at the apex and base, 0.20–2.00 µm thick (x̄=0.97 μm, n=25), and bearing a hyaline, apical mucoid appendage. Appendage 4.70–11.0 × 0.80–1.20 µm (x̄=7.37 × 0.98 µm, n=10), flexible, unbranched, straight to curved, tapering towards an acutely rounded tip.
Material examined – Thailand, Chiang Mai Province, Forests around the Mushroom Research Center, on dead leaves of Musa sp., 1st April 2021, D. Gomdola (MFLU XXX, holotype).
Distribution – Thailand (This study).
Notes – Phyllosticta chiangmaiensis, the novel taxon described herein, is sister to P. musaechinensis S.P. Wu, Z.Y. Liu & K.D. Hyde (GZAAS 6.1247T and GZAAS 6.1384) with strong support (100%ML, 100%MP, 1.00PP) (Figure 3). The characters of conidia match the species concept of Phyllosticta. Conidial length of P. chiangmaiensis ranges from 13.0–18.0 µm while that of P. musaechinensis (GZAAS 6.1247) ranges between 14.0–18.0 µm, which are nearly similar. However, the conidiomata diameters and sheath thickness of our new taxon are bigger than that of the closely related taxa (Table 6). Other differences and similarities between P. chiangmaiensis and sister taxa are given (Table 6). Despite the fact that our new taxon grouped with other species (P. musaechinensis, P. musarum, P. maculata, P. cavendishii) that were isolated from Musa sp., it formed distinct lineages. In pairwise nucleotide comparisons of the type species of P. musaechinensis and our new taxon P. chiangmaiensis, there are 6 nucleotide base pair (bp) differences in ITS (526 nucleotides), 16 bp differences in ef1α (165 nucleotides) and 1 bp differences in act (210 nucleotides). In ITS region of the type species of P. musarum, 13 nucleotide bp differences were observed across 431 nucleotides.
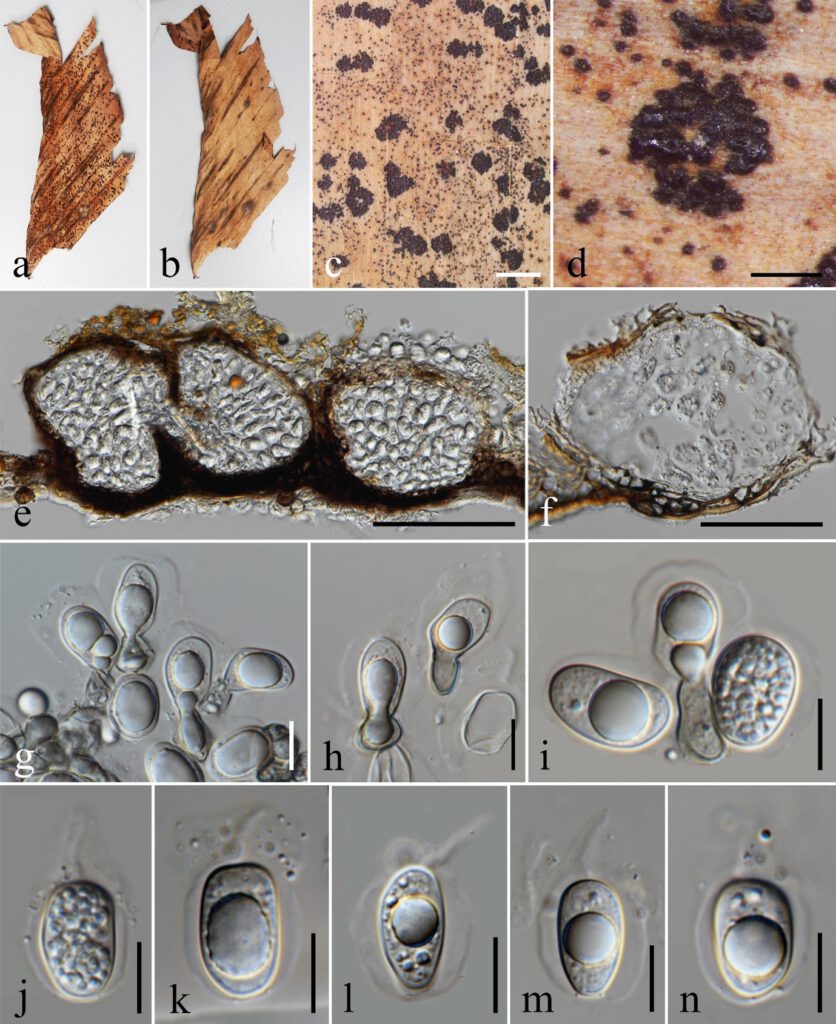
Figure 1 – Phyllosticta chiangmaiensis sp. nov. (holotype) a–b Appearance of conidiomata on leaves of Musa sp. c–d Close up of conidiomata on substrate. e–f Section through conidiomata showing pycnidial wall. g–i Conidiophores reduced to conidiogenous cells. j–n Conidia surrounded by mucilaginous sheath, bearing an apical appendage. Scale bars: c = 2 mm, d = 500 μm, e = 100 μm, f = 50 μm, g–n = 10 μm.
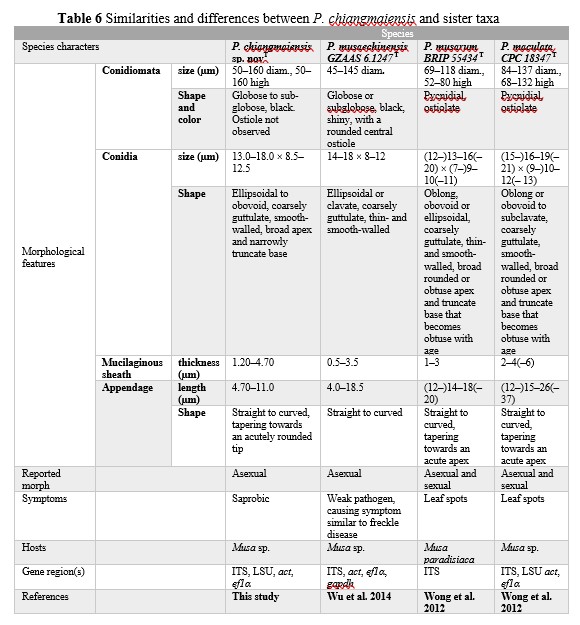
Genealogical Concordance Phylogenetic Species Recognition Analysis (GCPSR)
The pairwise homoplasy index resulted in a threshold exceeding 0.05 (Фw = 0.837) for our newly described taxon, P. chiangmaiensis, indicating no significant recombination in the dataset. This means that recombination levels are different between related species in the same clades that reside P. chiangmaiensis. If the PHI is lower than the 0.05 threshold (Фw < 0.05), it indicates that there is recombination, indicating that related species in a group level are same (Figures 2).
Figure 2 – PHI test result using both LogDet transformation and splits decomposition.

Figure 3 – Phylogram generated from maximum likelihood analysis (RAxML) based on the combined ITS, LSU, EF-1α, ACT, GAPDH and RPB2 matrices of Phyllosticta. Maximum likelihood (ML) with bootstrap support ≥60% is given at respective nodes. The tree is rooted with Botryosphaeria obtusa (CMW 8232 and CMW 7775) and B. stevensii (CBS 112553 and CMW 7060). Ex-type strains are indicated in bold and our isolates are in bold red.
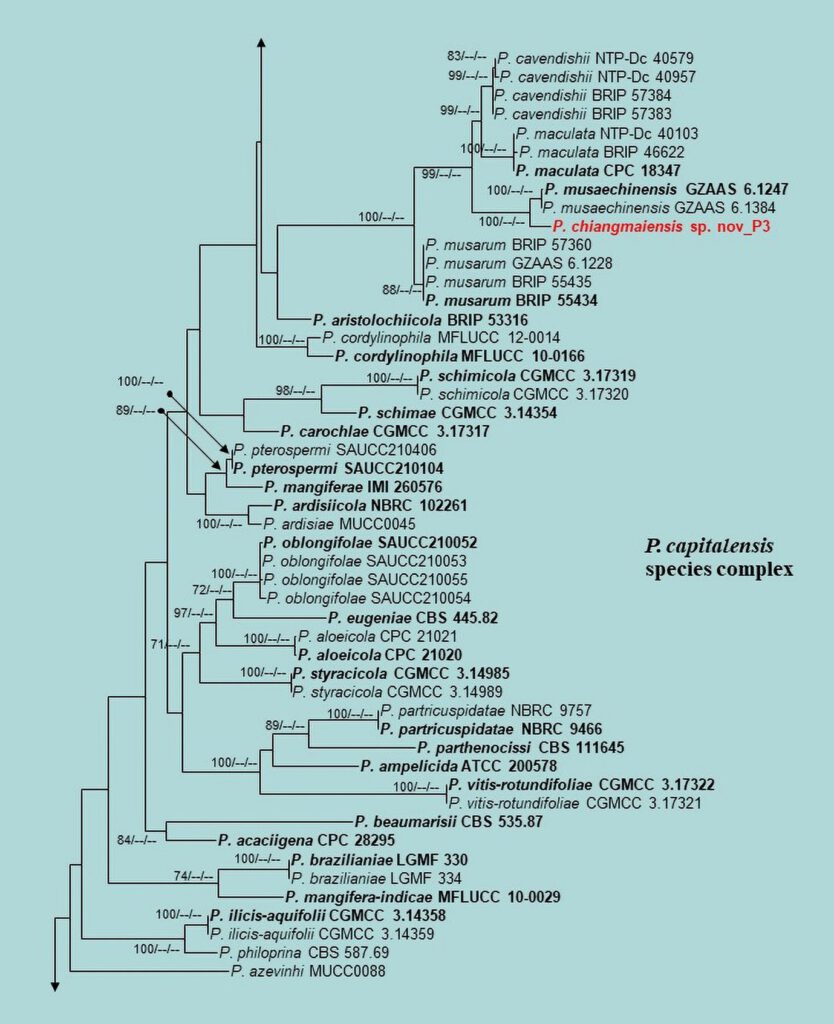
Figure 3 – Continued.

Figure 3 – Continued.
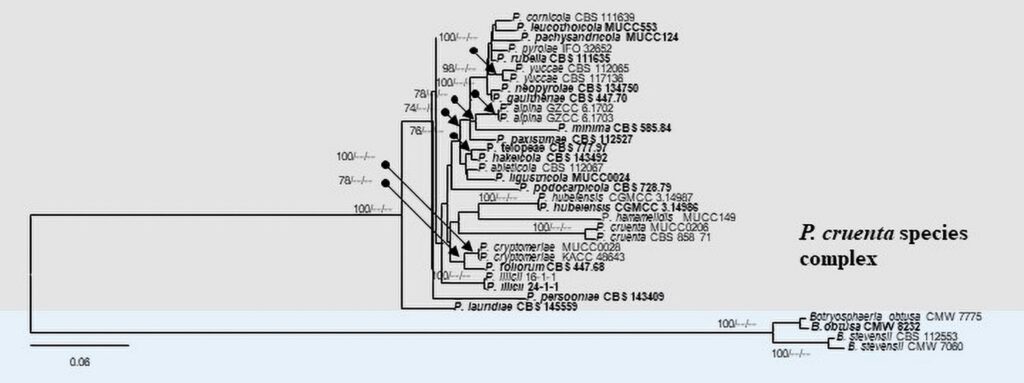
Figure 3 – Continued.
