Penicillium subrubescens Houbraken, Mansouri, Samson & Frisvad, Antonie van Leeuwenhoek 103(6): 1354 (2013).
MycoBank number: MB 801306; Index Fungorum number: IF 801306; Facesoffungi number: FoF 11349;
No synonyms
Description
Culture characteristics: Colonies growing after 5 days at 25 ± 2 ºC on following agar media: CYA colonies medium growing, floccose, sporulation turquoise white (24A2),18 mm in diam.; margins irregular; exudates colourless; soluble pigments absent; reverse wrinkled, butter yellow (4A5). MEA colonies medium growing, radially sulcate, velutinous, sporulation pale yellow (1A3), 22 mm in diam.; margins irregular; exudate absent; soluble pigment absent; reverse sulcate, butter yellow (4A5). CYAS colonies slow-growing, floccose, mycelium white (1A1), 9 mm in diam.; margin irregular; exudate absent; soluble pigment absent; reverse greyish yellow (4B6). OA colonies medium growing, flat, granular, mycelium white, 6 mm in diam.; margins irregular; sporulation yellowish-white (1A2); exudates colourless; soluble pigment absent; reverse yellowish-white (1A2). CZ colonies slow-growing, flat, granular, mycelium white (1A1), 13 mm in diam.; margins irregular; exudate absent; soluble pigment absent; reverse white (1A1). DG18 colonies slow-growing, flat, mycelium white (1A1), 6 mm in diam.; margin irregular; exudate absent; soluble pigment absent; reverse white (1A1). YES colonies medium growing, floccose, mycelium white (1A1), 21 mm in diam.; margins irregular; sporulation orange white (5A2); exudate absent; soluble pigment absent; reverse wrinkled, sulcate, butter yellow (4A5). CREA 25 °C, 5 d, in darkness: Colonies slow-growing, flat 10 mm in diam.; margin irregular; acid production absent; sporulation yellowish-white (3A2); reverse white (1A1). Conidiophores predominantly biverticillate, often with one or more additional branches that also have symmetrically biverticillate structures; stipes coarsely roughened, 150–400 × 2.5–4 μm; metulae in verticils of 3 to 6, walls roughened, both equal and unequal in length, 10–15 (−20) × 2.0– 3.5 μm; phialides ampulliform with distinct neck, with walls occasionally roughened, 7.0–9.5 × 2.0–3.5 μm; conidia subglobose to broadly ellipsoidal, smooth or finely roughened, 2.5–3.0 × 2.0–2.7 μm.
Material examined: INDIA, Uttarakhand, Almora (29°35′15″N, 79°35′46″E), from soil sample, 27 February 2019, Rajeshkumar K.C., Nikhil Ashtekar (NFCCI 5063).
Distribution: USA, Tanzania, Finland, and Canada
Sequence data from this collection: ITS: OK345037 (ITS5/ITS4); BenA: OL652649 (Bt2a/Bt2b); CaM: OM948804 (CMD5/CMD6); RPB2: OL652655 (fRPB2-5F/fRPB2-7CR)
Notes: The concatenated phylogenetic analyses of ITS, BenA, CaM, and RPB2 genes revealed that our isolate (NFCCI 5063) clusters with the type stain, P. subrubescens (CBS H-21029) with a bootstrap value of 100%. Further, both these strains demonstrate the same morphological characters such as predominant symmetrically biverticillate conidiophore with globose to ellipsoidal conidia with a smooth ornamentation. We recognize this isolate as P. subrubescens based on the morphology and phylogeny and this is the first record of P. subrubescens in India.
Importance of species to humans or ecosystem: There is no record of any threat to humans or ecosystem
Quarantine significance: No data available regarding quarantine. Fungi belonging to BS level I
Biochemical importance, chemical diversity or application: P. subrubescens is an efficient producer of Inulinase (Mansouri et al. 2013). This species produced high plant cell wall-degrading enzyme and its saccharification abilities were similar to that of the industrial species A. niger (Mäkelä et al., 2016). The genomic analysis of P. subrubescens revealed an expanded set of Carbohydrate Active enzyme (CAZyme)-encoding genes (Peng et al., 2017). Recently, a comparative exoproteome and transcriptome analysis of P. subrubescens cultures grown on wheat bran and sugar beet pulp and showed that P. subrubescens adapts its enzyme production to the available plant biomass and is a promising new fungal cell factory for the production of CAZymes (Dilokpimol et al., 2020).
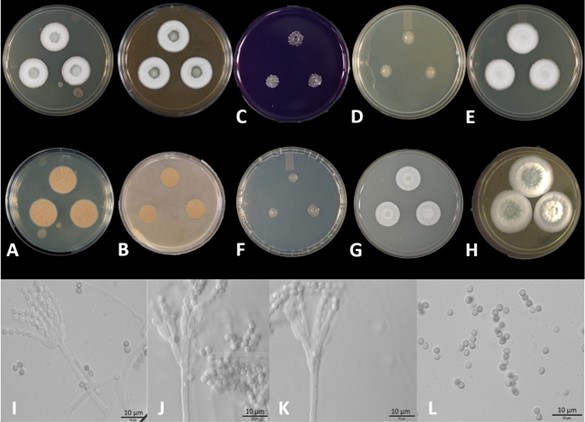
Fig. 1. Penicillium subrubescens. A, B Colonies after 5d at 25± 2 ºC on CYA and MEA obverse and reverse. C CREA obverse. D CYAS obverse. E CZA obverse. F DG18 obverse. G OA (natural) obverse. H YES obverse. I monoverticillate conidiophore. J-K biverticillate conidiophore. L Conidia. Scale bar: I-L=10 μm.
