Nigrospora lacticolonia Mei Wang & L. Cai, in Wang, Liu, Crous & Cai, Persoonia 39, 131 (2017)
Index Fungorum number: IF 820735; Facesoffungi number: FoF 141323; Fig. 1
Endophytic on Acrostichum aureum leaves. Sexual morph: Undetermined. Asexual morph: Hyphae 3–6 µm diam. (x̅ = 3.67 ± 0.83 μm, n = 30), septate, hyaline, smooth, branched. Conidiomata immersed, scattered, globose, black. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 6–12 × 5.5–9 μm (x̅ = 8.35 × 6.75 μm, n = 30), aggregated in clusters on hyphae, globose to clavate to bean-shaped, pale brown, verrucous. Conidia spherical diameter 11–15 μm (x̅ = 13.38 ± 1.03 μm, n = 30), elliptic 12.5–16 × 9–14 μm (x̅ = 14.13 ± 0.85 × 11.56 ± 1.22 μm, n = 30), solitary, spherical or slightly elliptical, black, shiny, smooth, aseptate.
Culture characteristics – Colonies on PDA reaching 8 cm diam. after 3 days in the dark at 25℃, circular, woolly in the center, darker in the center, the mycelium covering becomes thinner towards the edge, flocculent, surface view white, reverse view white to off-white, without pigmentation.
Material examined – China, Guangdong Province, Guangzhou city, from leaves of Acrostichum aureum, 15 September 2021, Li Hua (MHZU 23-0006, dried culture), living culture ZHKUCC 23-0023, ZHKUCC 23-0024, ZHKUCC 23-0025.
GenBank accession numbers – ZHKUCC 23-0023 – ITS: OQ799024, tef1-α: OQ858593, β-tubulin: OQ858596; ZHKUCC 23-0024 – ITS: OQ799025, tef1-α: OQ858594, β-tubulin: OQ858597; ZHKUCC 23-0025 – ITS: OQ799026, tef1-α: OQ858595, β-tubulin: OQ858598.
Known distribution (based on molecular data) – China, Jiangxi Province (Wang et al. 2017, Kee et al. 2019), Hainan Province (Wang et al. 2017), Guangxi Province (Raza et al. 2019), Guangdong Province (this study).
Known hosts (based on molecular data) – Acrostichum aureum (this study), Camellia sinensis (Wang et al. 2017), Hylocereus polyrhizus (Kee et al. 2019), Musa paradisiaca (Wang et al. 2017), Saccharum officinarum (Raza et al. 2019).
Notes – Nigrospora lacticolonia was introduced by Wang et al. (2017) from Jiangxi Province, China on Camellia sinensis leaves. In the present study, we isolated three Nigrospora strains from Acrostichum aureum leaves. Based on the BLASTn results from the NCBI database, N. lacticolonia was given as the closest match. In the multigene phylogenetic analysis of the concatenated ITS, β-tubulin, and tef1-α, our isolates (ZHKUCC 23-0023, ZHKUCC 23-0024, and ZHKUCC 23-0025) cluster with N. lacticolonia strains with 100% maximum likelihood support and 1.00 Bayesian posterior probability (Fig. 2). The morphology of our isolates is similar to that of the ex-type of N. lacticolonia. Based on multigene phylogeny and morphological description, we provide a new host record for N. lacticolonia from Acrostichum aureum, collected from Jiangxi Province, China.
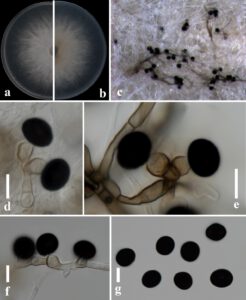
Fig. 1 – Nigrospora lacticolonia (ZHKUCC 23-0023, a new host record). a Colony on PDA from front view. b Colony on PDA from reverse view. c Conidial mass forming on the culture. d–f Conidia attached to conidiogenous cells. g Conidia. Scale bars: d–g = 10 μm.

Fig. 2 – Phylogram generated from maximum likelihood analysis based on the combined ITS, β-tubulin and tef-1α sequence data of Nigrospora. Ninety-three strains of Nigrospora are included in the combined analyses, which comprised 1164 characters (444 characters for ITS, 384 characters for β-tubulin, 334 characters for tef-1α). The RAxML analysis yielded the best scoring tree, which was used as the backbone tree, with a final likelihood value of -8177.485492. The matrix had 526 distinct alignment patterns with 7.86% of undetermined characters or gaps. Under the Akaike Information Criterion (AIC) the evolutionary model HKY+G is applied to ITS and β-tubulin genes, and GTR+I+G is applied to tef-1α gene. Maximum likelihood (ML) with bootstrap support greater than or equal to 50% and Bayesian posterior probabilities (PP) values greater than or equal to 0.8 are given near nodes, respectively. The tree is rooted with Apiospora vietnamensis (IMI 99670) and A. sichuanensis (HKAS 107008). Ex-type strains are in bold and the species described herein are denoted in yellow. Hyphen (-) represents support values below 50% (ML) and below 0.80 (PP).
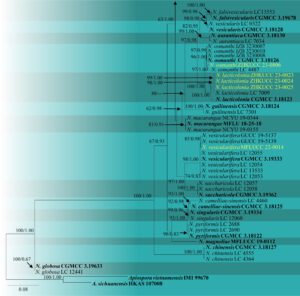
Fig. 2 – Continued.
