Neoschizothecium S.K. Huang & K.D. Hyde, gen. nov.
Etymology – Name refers to genus similar to, but different from Schizothecium
Index Fungorum number: IF558329; Facesoffungi number: FoF 10034
Saprobic on wood, coprophilous.
Sexual morph: Ascomata perithecial, solitary, scattered or gregarious, superficial to semi-immersed, pyriform, olivaceous-brown to dark brown, semi-transparent, surrounded by pale brown hyphae, ostiolate, with papilla. Peridium comprising membranaceous, pale brown cells of textura angularis. Paraphyses cylindrical, septate. Asci 8-spored, unitunicate, cylindrical, evanescent. Ascospores uniseriate to irregularly arranged, ellipsoidal to broadly fusiform, aseptate, hyaline to dark brown, truncate at the base with a cylindrical, slender, hyaline pedicel, umbonate apex, verrucose, with an apical germ pore.
Asexual morph: Undetermined (adapted from Corda 1838; Lundqvist 1972; Munngai et al. 2012; Wang et al. 2019a, b).
Notes – Schizothecium was introduced by Corda (1838) and is typified by S. fimicola and has membranaceous asco- mata and ascospores with lash-like caudae (Corda 1838; Lundqvist 1972; Hu et al. 2006; Mungai et al. 2012). The generic type, Schizothecium fimicola, was synonymized as Podospora fimicola based on the epitype strain S. fimicola (CBS 482.64), which nested in Podospora (Podosporaceae) (Wang et al. 2019a, b; Index Fungorum 2020). Marin-Felix et al. (2020) established Schizotheciaceae and raised Schizothecium as its type genus; however, the generic type of Schizothecium has not been clarified. Currently, Schizothecium is a synonym of Podospora under Podosporaceae, whereas several Schizothecium species clustered in Schizotheciaceae (Marin-Felix et al. 2020; this study, 100%ML/1.00PP, Fig. 26). In this study, a novel genus Neoschizothecium is therefore proposed to accommodate species in the Neoschizotheciaceae clade (Fig. 26) and is typified by Neo. curvisporum (see the combination list below). This genus nests in Neoschizotheciaceae (95%ML) and is distinct from Podospora in multi-gene analysis (Fig. 26).
Type species – Neoschizothecium curvisporum (Cain) S.K. Huang & K.D. Hyde, comb. nov.
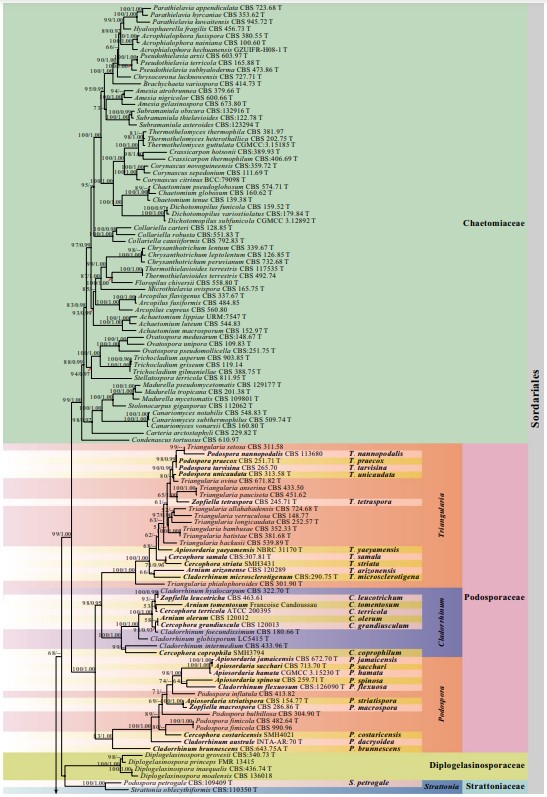
Fig. 26 Phylogram generated from maximum likelihood analysis based on combined LSU, ITS, TUB and RPB2 sequence data of Sordariales. The confdence values of bootstrap (BS) proportions from the Maximum Likelihood (ML) analysis (MLBS>50%, before the backslash) and the posterior probabilities (PP) from the Bayesian (BY) analysis (BY-PP>0.90, after the backslash) above corresponding nodes. The ‘–’ indicates lack of statistical support (<50% for ML-BS and<0.90 for BY-PP). Two hundred and thirty-fve strains are included in the combined analyses, which comprise 2877 characters (851 characters for LSU, 509 characters for ITS, 665 characters for TUB, 852 characters for RPB2) after alignment. Strains of Microascales are used as outgroup taxa. The best score in IQ-TREE explores with a fnal likelihood value of − 83459.1151 is presented. The model of each partitioned gene is: LSU: GTR+F+I+G4; ITS: TIM2e+I+G4; TUB: HKY+F+I+G4; RPB2: TIM3+F+I+G4. Strain numbers are noted after the species names and ex-type strains are marked with ‘T’ after the culture number. Alignments are available at TreeBASE (URL: http://purl.org/phylo/treebase/ phylows/study/TB2:S28270)
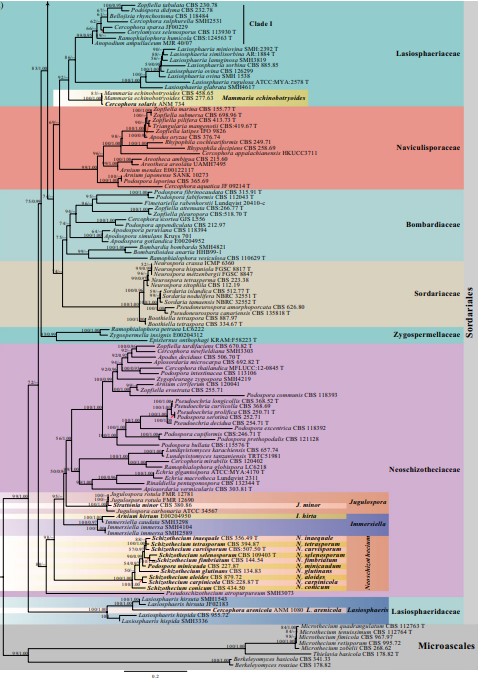
Fig. 26 (continued)
Species
