Neoschizothecium curvisporum (Cain) S.K. Huang & K.D. Hyde, comb. nov.
Basionym: Sordaria curvispora Cain, Canadian Journal of Research, Section C 26: 492 (1948)
Synonym: Schizothecium curvisporum (Cain) N. Lundq., Symb. bot. upsal. 20(no. 1): 334 (1972)
MycoBank number: MB 558333; Index Fungorum number: IF 558333; Facesoffungi number: FoF 10136; Fig. 52q–r
Saprobic on seeds. Sexual morph: Ascomata perithecial, gregarious, superficial, pyriform, olivaceous brown to dark brown, semi-transparent, surrounded by pale brown hyphae, ostiolate, with papilla, periphysate. Peridium composed of membranaceous, pale brown cells of textura angularis. Asci 8-spored, unitunicate, cylindrical, apex rounded, pedicellate. Ascospores uni- or bi-seriate, reniform, aseptate, brown to dark brown, truncate at the base with a clavate, hyaline pedicel, with an apical germ pore. Asexual morph: Undetermined.
Known hosts and distribution: On seeds of Apium graveolens and Daucus carota in the USA (type locality) (Cain 1948).
Notes: Schizothecium curvisporum was established based on Sordaria curvispora which was isolated in culture from seed of Daucus carota L. var. sativa DC. in the USA (DAOM 20504) (Cain 1948; Lundqvist 1972). Subsequently, Vu et al. (2019) sequenced the ex-type strain S. curvisporum (CBS 506.50), and this strain nests in Neoschizothecium (88%ML, Fig. 26). In this study, we were unable to obtain authentic materials. Ascomata of N. curvisporum is similar to N. conicum and the hand-drawing is provided for the asci and ascospores (Fig. 52q–r) based on Cain (1948).
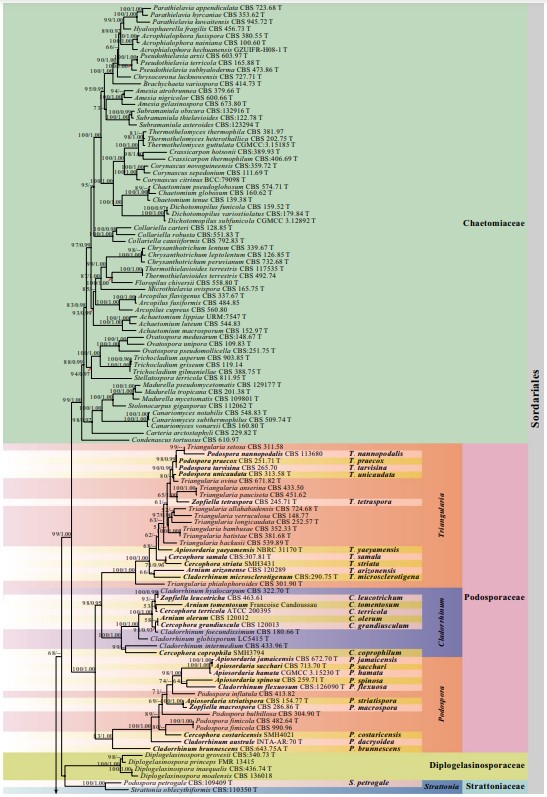
Fig. 26 Phylogram generated from maximum likelihood analysis based on combined LSU, ITS, TUB and RPB2 sequence data of Sordariales. The confdence values of bootstrap (BS) proportions from the Maximum Likelihood (ML) analysis (MLBS>50%, before the backslash) and the posterior probabilities (PP) from the Bayesian (BY) analysis (BY-PP>0.90, after the backslash) above corresponding nodes. The ‘–’ indicates lack of statistical support (<50% for ML-BS and<0.90 for BY-PP). Two hundred and thirty-fve strains are included in the combined analyses, which comprise 2877 characters (851 characters for LSU, 509 characters for ITS, 665 characters for TUB, 852 characters for RPB2) after alignment. Strains of Microascales are used as outgroup taxa. The best score in IQ-TREE explores with a fnal likelihood value of − 83459.1151 is presented. The model of each partitioned gene is: LSU: GTR+F+I+G4; ITS: TIM2e+I+G4; TUB: HKY+F+I+G4; RPB2: TIM3+F+I+G4. Strain numbers are noted after the species names and ex-type strains are marked with ‘T’ after the culture number. Alignments are available at TreeBASE (URL: http://purl.org/phylo/treebase/ phylows/study/TB2:S28270)
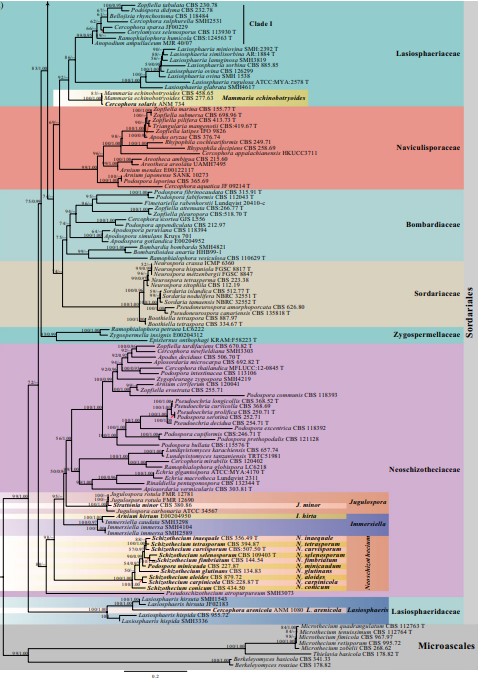
Fig. 26 (continued)

Fig. 52 Neoschizothecium conicum: a, b, e, h–l, p (F124745); c, d, f, g, m, n (S-F124742); o (redrawn from Fuckel 1870). a Herbarium material. b, c Ascoma on host. d Ascoma in cross section. e, f Peridium. g, h Wall structure around ostiole with periphyses. i–k Asci. l–p Ascospores; Neoschizothecium curvisporum: q, r (redrawn from Cain 1948). q Asci. r Ascospores. Scale bars: b. c = 500 µm, d = 200 µm, e–g. l–p = 20 µm, h–k = 50 µm, q. r = 10 µm
