Neogloeosporidina W.J. Li, Camporesi & K.D. Hyde, gen. nov.
Index Fungorum number: IF557154, Facesoffungi number: FoF 07519
Etymology: Named after its morphology similar to Gloeosporidina.
Saprobic on plant host in terrestrial habitat. Sexual morph: undetermined. Asexual morph: Conidiomata brown, eustromatic, pycnidial, scattered, rarely gregarious, subepidermal or subperidermal in origin, immersed to partly erumpent, globose to subglobose, unilocular, thick-walled, smooth, glabrous. Ostiole absent, dehiscing by an irregular rupture in the apical region, then becoming wide open. Conidiomatal wall composed of brown, thick-walled cells of compressed textura angularis. Conidiophores mostly reduced to conidiogenous cells, occasionally present, short, cylindrical, septate and branched at the base, formed from inner layers of conidiomata. Conidiogenous cells hyaline, entero-blastic, phialidic, cylindrical, usually integrated, determinate, smooth-walled. Conidia hyaline, ellipsoid to ventricose, often with a blunt apiculus, unicellular, thick- and smooth-walled.
Type species: Neogloeosporidina pruni W.J. Li, Campo- resi & K.D. Hyde, sp. nov.
Notes: Neogloeosporidina pruni was collected from Italy. The sequence was obtained directly from conidiomata. Multi-locus phylogeny reveals N. pruni clusters in a well-supported branch basal to Dermea and Corniculariella (Fig. 129). Neogloeosporidina pruni shares similar characters with Gloeosporidina in having elliptical conidia and enteroblastic, phialidic, integrated conidiogenous cells, but it can be distinguished by its eustromatic, pycnidial conidi- omata. Combining morphology and phylogeny, Neo- gloeosporidina is introduced as a new genus.
Distribution: Italy.
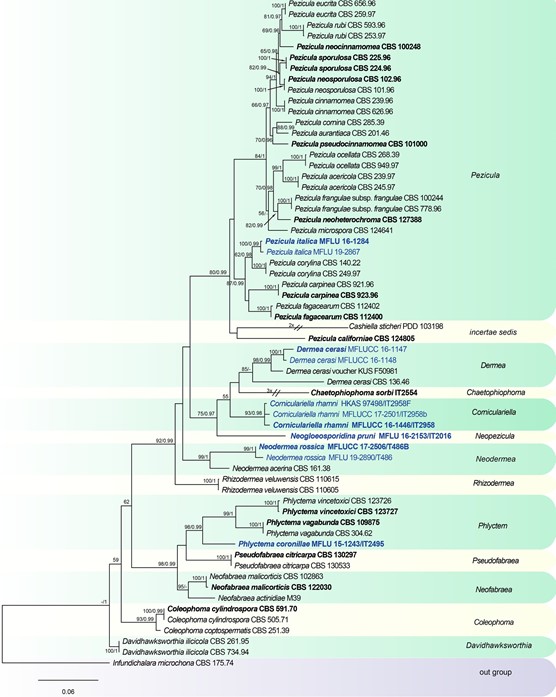
Fig. 129 Phylogenetic tree generated from a maximum likelihood analysis based on a concatenated alignment of LSU, ITS and rpb2 sequences data of Dermateaceae. The newly generated nucleotide sequences were compared against the GenBank database via blast search. Related sequences were obtained from (Chen et al. 2016) and GenBank. Sixty-two strains are included in the analyses, which comprise 2465 characters including gaps. Infundichalara microchona CBS 175.74 is used as the outgroup taxon. The tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best scoring RAxML tree with a final optimization likelihood value of − 15611.979066 is presented. The matrix had 938 distinct alignment patterns, with 16.00% of undetermined characters or gaps. Esti- mated base frequencies were: as follows A = 0.251579, C = 0.230991, G = 0.272796, T = 0.244634; substitution rates AC = 1.650679, AG = 3.597104, AT = 0.907930, CG = 0.800954, CT = 8.324139, GT = 1.000000; gamma distribution shape parameter α = 0.201567. Maximum likelihood (MLBS) bootstrap support values higher than 50%, and Bayesian posterior probabilities ≥ 0.95 (PP) are shown above or below the nodes. Hyphen (“–”) indicates a value lower than 50% for MPBS and MLBS and a posterior probability lower than 0.95 for BYY. The scale bar indicates 0.06 changes. Ex-type or ex- epitype strains are in bold. New isolates are in blue
Species
