Longiseptatispora curvata Crous & Bulgakov, sp. nov.
MycoBank number: MB 835084; Index Fungorum number: IF 835084; Facesoffungi number: FoF 12271; Fig. 32.
Etymology: Name refers to the curved conidia of this species.
Conidiomata pycnidial, brown, globose, immersed to erumpent, 250–300 µm diam, with 1–2 ostioles; wall of 3–6 layers of brown textura angularis. Conidiophores reduced to conidiogenous cells (or with a supporting cell), lining the inner cavity, doliiform to ampulliform, 5–8 × 5–7 µm, phialidic with percurrent proliferation. Conidia hyaline, smooth, granular, solitary, subcylindrical with obtuse apex and truncate base, straight to irregularly curved, (0–)1-septate, (20–)25–32(–37) × 4(–5) µm.
Culture characteristics: Colonies flat, spreading, with moderate aerial mycelium and smooth, lobate margin, reaching 60 mm diam after 2 wk at 25 ºC. On MEA surface dirty white, reverse buff; on PDA surface dirty white in centre, grey olivaceous in outer region, reverse iron-grey; on OA surface dirty white in centre, grey olivaceous in outer region.
Typus: Russia, Rostov region, Shakhty city district, trees near Atyukhta river, on living and dying twigs of Lonicera tatarica, 22 Sep. 2018, T.S. Bulgakov, HPC 2616 = Myc-22 (holotype CBS H-24166, culture ex-type CPC 36457 = CBS 146035).
Additional material examined: Russia, Rostov region, Shakhty city district, Alexandrovsky park, on live and dying twigs of Lonicera tatarica (Caprifoliaceae), 3 Nov. 2018, T.S. Bulgakov, HPC 2684 = Myc-71, culture CPC 36771.
Notes: The DNA phylogeny (Fig. 33) showed that the two strains from Russia on Lonicera tatarica formed a fully supported clade. This was also true for an isolate collected from the Netherlands on Melilotus sp. with a sequence from GenBank (accession AF439466.1) deposited under the name Leptosphaeria weimeri. The association between these two lineages is not well-supported (PP = 0.81/ML-BS = 75 %), but they are clearly different from the other known genera in Leptosphaeriaceae. Therefore the new genus Longiseptatispora is proposed here to accommodate these taxa. A greater number of isolates and phylogenetically informative gene sequences are needed to determine whether these two lineages might represent distinct genera. Morphologically, Lo. curvata shares some characters with the other species (Lo. meliloti) in this genus by producing long, septate conidia, but could be differentiated from the latter by its longer and less septate conidia: conidia of Lo. curvata are (0–)1-septate, (20–)25–32(–37) × 4(–5) µm, while conidia of Lo. meliloti are (0–)3-septate, 15–26 × 3.5–5.5 µm. In addition, Lo. curvata also differs from the latter species by producing pycnidia with 1–2 ostioles while pycnidia of Lo. meliloti have a single central ostiole.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence of CPC 36457 had highest similarity to Paraleptosphaeria dryadis (strain CBS 643.86, GenBank MH862007.1; Identities = 546/578 (94 %), 8 gaps (1 %)), Coniothyrium wernsdorffiae (strain CBS 150.34, GenBank MH855474.1; Identities = 546/579 (94 %), 9 gaps (1 %)), and Phoma schachtii (strain CBS 502.84, GenBank MH861770.1; Identities = 541/578 (94 %), 7 gaps (1 %)). The ITS sequences of CPC 36457 and 36771 are identical (573/573 bases) – also see Fig. 33. Closest hits using the LSU sequence of CPC 36457 are Subplenodomus galicola (voucher MFLU 15-1368, GenBank KY554199.1; Identities = 836/842 (99 %), no gaps), Subplenodomus iridicola (strain CBS 143395, GenBank MH107965.1; Identities = 825/832 (99 %), no gaps), and Subplenodomus violicola (strain CBS 306.68, GenBank GU238156.1; Identities = 825/832 (99 %), no gaps) – also see Fig. 3, part 3 and Fig. 33. The LSU sequences of CPC 36457 and 36771 are identical (815/815 bases). Closest hits using the rpb2 sequence of CPC 36457 had highest similarity to Neopyrenochaeta cercidis (as Neopyrenochaeta sp. SCJ-2019a; voucher MFLU 18-2089, GenBank MK434908.1; Identities = 645/775 (83 %), 2 gaps (0 %)), Curvularia hawaiiensis (strain CBS 727.96, GenBank HG779168.1; Identities = 580/697 (83 %), 8 gaps (1 %)), and Plenodomus hendersoniae (strain LTO, GenBank MF795832.1; Identities = 602/725 (83 %), 8 gaps (1 %)). Closest hits using the tef1 (second part) sequence of CPC 36457 had highest similarity to Stemphylium callistephi (strain EEB 1055, GenBank JQ672393.1; Identities = 403/418 (96 %), no gaps), Wojnowiciella dactylidis (strain CBS 145077, GenBank MK442724.1; Identities = 395/410 (96 %), no gaps), and Paraphoma radicina (strain UTHSC DI16-209, GenBank LT797075.1; Identities = 395/410 (96 %), no gaps). The tef1 sequences of CPC 36457 and 36771 are identical (418/418 bases). Closest hits using the tub2 sequence of CPC 36457 had highest similarity to Parafenestella salicis (strain C303, GenBank MK357628.1; Identities = 450/520 (87 %), 19 gaps (3 %)), Parafenestella pseudosalicis (strain C301, GenBank MK357620.1; Identities = 445/514 (87 %), 19 gaps (3 %)), and Parafenestella pseudoplatani (strain C26, GenBank MF795914.1; Identities = 444/515 (86%), 19 gaps (3 %)). The tub2 sequences of CPC 36457 and 36771 are identical (535/535 bases).
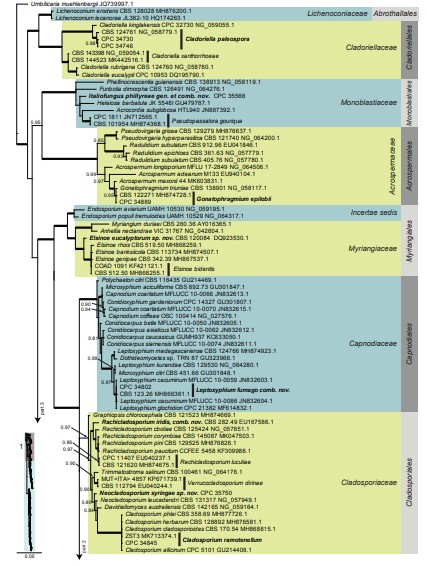
Fig. 3, parts 1–4. Consensus phylogram (50 % majority rule) resulting from a Bayesian analysis of the Dothideomycetes LSU sequence alignment. Bayesian posterior probabilities (PP) > 0.79 are shown at the nodes and the scale bar represents the expected changes per site. Thickened branches represent PP = 1. Families and orders are indicated with coloured blocks to the right of the tree. GenBank accession and/or culture collection numbers are indicated for all species. The tree was rooted to Umbilicaria muehlenbergii (strain A18, GenBank JQ739997.1) and the species treated in this study for which LSU sequence data were available are indicated in bold face. A miniature overview tree is also presented to facilitate navigation along the tree topology.
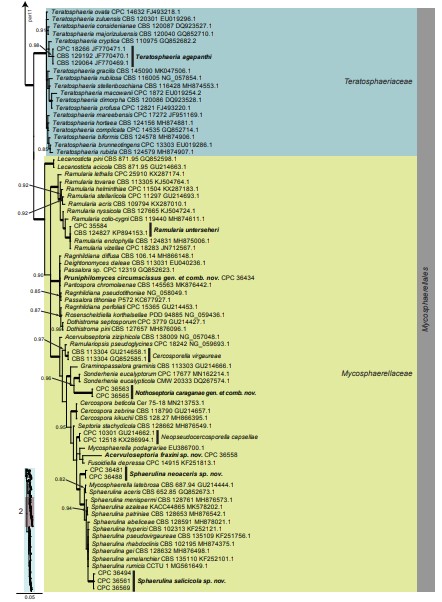
Fig. 3. (Continued).
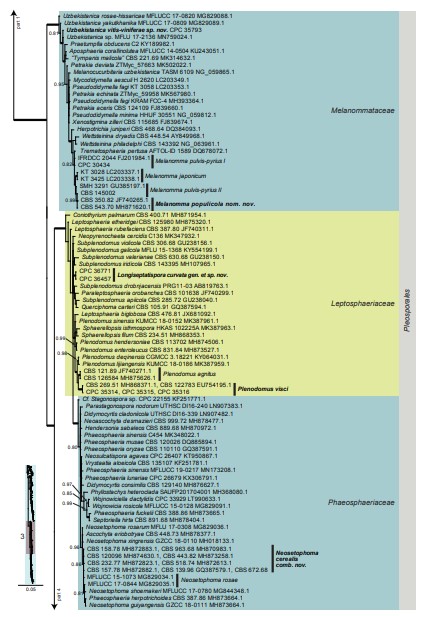
Fig. 3. (Continued).
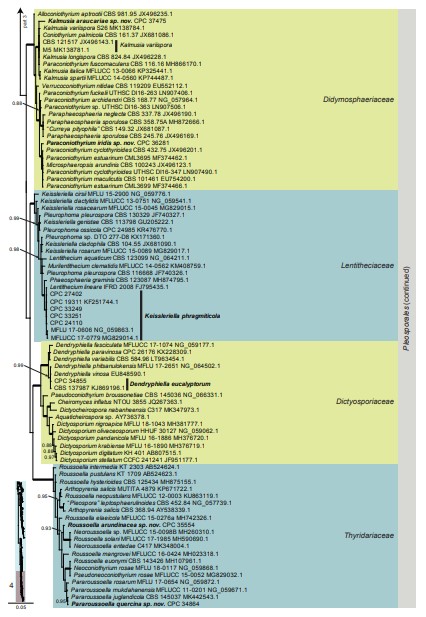
Fig. 3. (Continued).
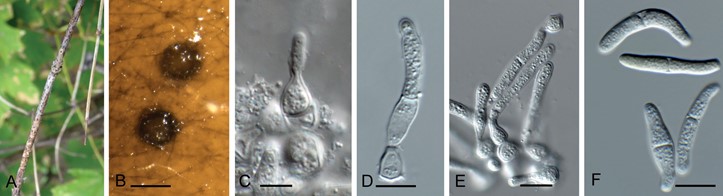
Fig. 32. Longiseptatispora curvata (CPC 36457). A. Conidiomata on stems of Lonicera tatarica. B. Conidiomata on MEA. C–E. Conidiogenous cells giving rise to conidia. F. Conidia. Scale bars: B = 300 µm, all others = 10 µm.
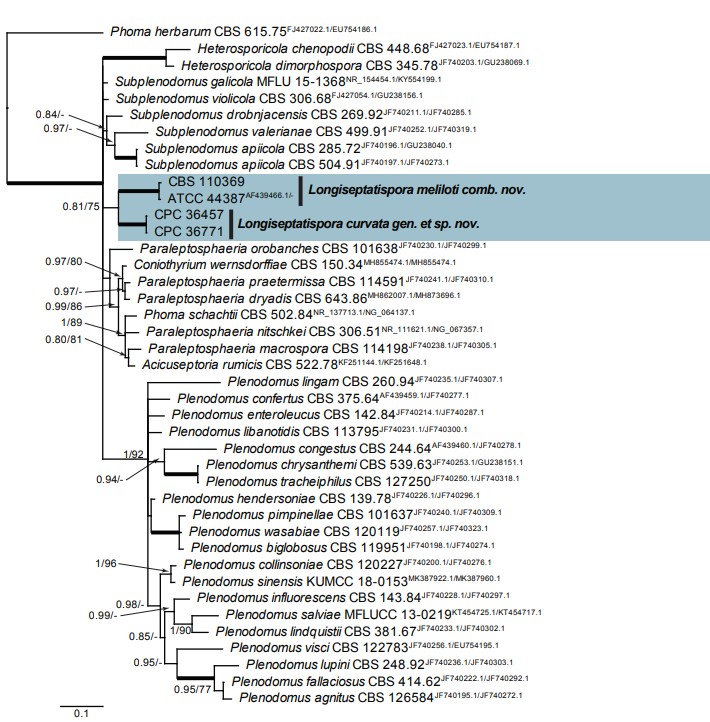
Fig. 33. Consensus phylogram (50 % majority rule) resulting from a Bayesian analysis of the Leptosphaeriaceae ITS/LSU sequence alignment (41 strains including the outgroup; 83 and 228 unique site patterns for ITS and LSU, respectively; 275 252 sampled trees from 1 835 000 generations). The alignment is derived from the ITS/LSU alignment of De Gruyter et al. (2013). Bayesian posterior probabilities (PP) > 0.79 and Maximum likelihood (as implemented in MEGA v. 7, Kumar et al. 2016) bootstrap support values (ML-BS) > 74 % are shown at the nodes (PP/ML-BS) and the scale bar represents the expected changes per site. Thickened branches represent PP = 1. GenBank accession (superscript text) and/or culture collection numbers are indicated for all species. The tree was rooted to Phoma herbarum (culture CBS 615.75; GenBank FJ427022.1 and EU754186.1 for ITS and LSU, respectively) and the species treated in this study are indicated in bold face.
