Italiofungus phillyreae (Thüm.) Crous, comb. nov.
MycoBank number: MB 835079; Index Fungorum number: IF 835079; Facesoffungi number: FoF 11956; Fig. 26.
Basionym: Depazea phillyreae Thüm., Boll. Soc. Adriatica Sci. Nat. Triests 2: 455. 1877.
Synonym: Phyllosticta goritiensis Sacc., Syll. Fung. (Abellini) 3: 22. 1884.
Conidiomata pycnidial, solitary, globose, brown with central ostiole, 200–300 µm diam; wall of several layers of brown textura intricata. Conidiophores arising from inner layer, hyaline, smooth, septate, branched, subcylindrical, straight to geniculate-sinuous, 10–30 × 2–3 µm. Conidiogenous cells integrated, terminal and intercalary, hyaline, smooth, subcylindrical with apical taper, proliferating sympodially at apex, scars not thickened nor darkened, 4–12 × 1.5–2 µm. Conidia solitary, hyaline, smooth, aseptate, subcylindrical, straight, apex obtuse, base truncate, (3.5–)4–5 × (1.5–)2 µm.
Culture characteristics: Colonies flat, spreading, surface folded with sparse aerial mycelium and smooth, lobate margin, reaching 30 mm diam after 2 wk at 25 ºC. On MEA, PDA and OA surface olivaceous grey, reverse iron-grey.
Material examined: Italy, Rome, on leaves Phillyrea latifolia (Oleaceae), 13 Apr. 2018, P.W. Crous, HPC 2336 (CBS H-23937, culture CPC 35566 =CBS 145537).
Notes: The oldest name that applies to the present collection is Depazea phillyreae, a microconidial morph described from leaves of Phillyrea in Italy. The genus Heleiosa (based on H. barbatula) applies to a bitunicate ascomycete described from leaves of Juncus roemerianus collected in North Carolina (Kohlmeyer et al. 1996), which is closely related to, but distinct from the new genus occurring on Phillyrea in Italy.
Based on a megablast search of NCBI’s GenBank nucleotide database, only distant hits were obtained using the ITS sequence, such as Didymosphaeria futilis (strain CMW 22186, GenBank EU552123.1; Identities = 482/546 (88 %), 21 gaps (3 %)), Pseudopassalora gouriqua (strain CBS 101954, GenBank NR_160207.1; Identities = 451/520 (87 %), 25 gaps (4 %)), and Funbolia dimorpha (strain CPC 14170, GenBank JF951136.1; Identities = 468/561 (83 %), 29 gaps (5 %)). Closest hits using the LSU sequence are Pseudopassalora gouriqua (strain CBS 101954, GenBank MH874368.1; Identities = 821/851 (96 %), 2 gaps (0 %)), Heleiosa barbatula (strain JK 5548I, GenBank GU479787.1; Identities = 814/853 (95 %), 5 gaps (0 %)), and Funbolia dimorpha (strain CPC 14170, GenBank JF951156.1; Identities = 806/851 (95 %), 3 gaps (0 %)) – also see Fig. 3, part 1.
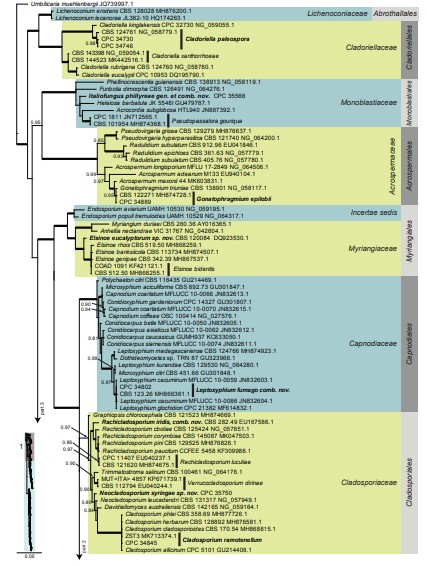
Fig. 3, parts 1–4. Consensus phylogram (50 % majority rule) resulting from a Bayesian analysis of the Dothideomycetes LSU sequence alignment. Bayesian posterior probabilities (PP) > 0.79 are shown at the nodes and the scale bar represents the expected changes per site. Thickened branches represent PP = 1. Families and orders are indicated with coloured blocks to the right of the tree. GenBank accession and/or culture collection numbers are indicated for all species. The tree was rooted to Umbilicaria muehlenbergii (strain A18, GenBank JQ739997.1) and the species treated in this study for which LSU sequence data were available are indicated in bold face. A miniature overview tree is also presented to facilitate navigation along the tree topology.
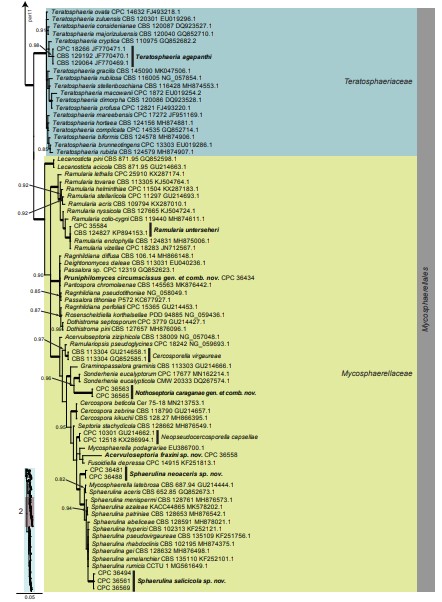
Fig. 3. (Continued).
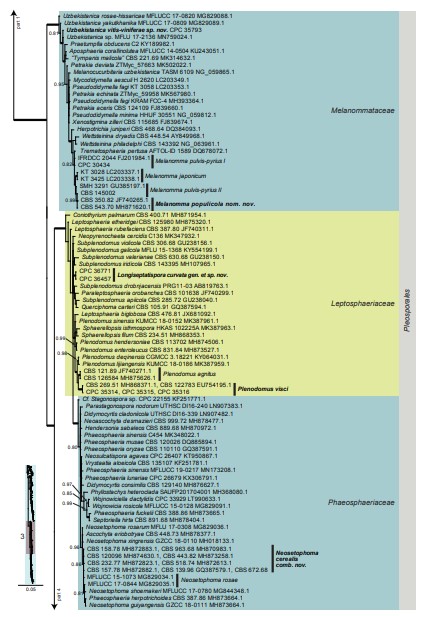
Fig. 3. (Continued).
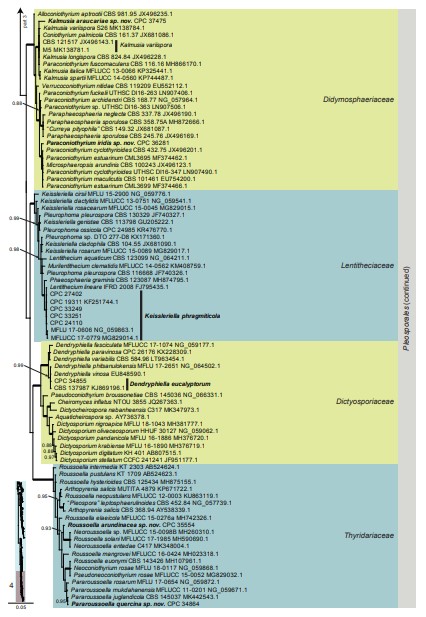
Fig. 3. (Continued).
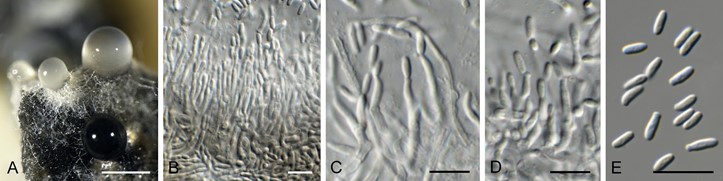
Fig. 26. Italiofungus phillyreae (CPC 35566). A. Conidiomata on PDA. B–D. Conidiophores with conidiogenous cells. E. Conidia. Scale bars: A = 300 µm, all others = 10 µm.
