Neolamproconium silvestre Crous & Akulov, sp. nov.
MycoBank number: MB 835090; Index Fungorum number: IF 835090; Facesoffungi number: FoF; Fig. 38.
Etymology: Name refers to its location, collected in a forest.
In vivo: Ascomata immersed in host tissue, perithecial, black, erect to oblique, erumpent, in clusters of 6–15, long necked with central ostioles, converging through a black stromatic disc, 1–1.5 mm diam; ascomata subglobose, in diatrypoid configuration, up to 350 µm diam, neck up to 600 µm long, slightly swollen at the tips; ostiole with hyaline periphyses; peridium of thick-walled cells of brown textura angularis; hamathecium of hyaline, septate, branched, hyphae-like paraphyses, 2.5–5 µm diam. Asci 8-spored, subcylindrical with obtuse to slightly flattened apex and apical mechanism (not bluing in Meltzer’s reagent), stipitate, arranged in basal layer, 90–140 × 13–16 µm. Ascospores hyaline, smooth, guttulate, granular, fusoid to subcylindrical, ends subobtuse, slightly constricted at median septum, mostly straight, (27–) 30–35(–38) × (6–)7(–8) µm (in water). Conidiomata erumpent through bark, black, stromatic, with aggregated rosettes of up to 15 pycnidia, upper part with slightly elongated black necks. In vitro: Conidiomata erumpent, solitary, pycnidial, globose, 200–300 µm diam, with central ostiole; wall of 3–6 layers of pale brown textura angularis. Conidiophores lining inner layer, hyaline, smooth, septate, branched, 40–80 × 2.5–4 µm. Conidiogenous cells hyaline, smooth-walled, subcylindrical, terminal and intercalary, 15–25 × 2–2.5 µm, phialidic with prominent periclinal thickening. Conidia dimorphic. Macroconidia hyaline, smooth, guttulate, granular, fusoid, apex subobtuse, base truncate, 1(–3)-septate, (28–)34–38(–42) × 4(–4.5) µm. Microconidia developing in same conidioma as macroconidia on SNA, rod-shaped, ends obtuse, curved to straight, aseptate, (6–)7–8 × 1.5(–2) µm.
Culture characteristics: Colonies flat, spreading, with sparse to moderate aerial mycelium and smooth, lobate margin, reaching 40 mm diam after 2 wk at 25 ºC. On MEA surface olivaceous grey with patches of honey, reverse isabelline to cinnamon; on PDA surface and reverse olivaceous grey; on OA surface ochreous.
Typus: Ukraine, Kharkiv, Forest-park, Sokol`niki-Pomerki, on branch of Tilia sp. (Malvaceae), 8 Oct. 2017, A. Akulov, HPC 2274 = CWU (Myc) AS 6601 (holotype specimen CBS 145566, preserved as metabolically inactive culture, culture ex-type CPC 34865 = CBS 145566).
Additional material examined: Ukraine, Kharkiv, Forest-park, Sokol`niki- Pomerki, on branch of Quercus robur (Fagaceae), 8 Oct. 2017, A. Akulov, HPC 2273 = CWU (Myc) AS 6660, culture CPC 34862.
Notes: Lamproconium, based on L. desmazieri, was recently treated by Norphanphoun et al. (2016), based on specimens on dead branches of Tilia cordata collected in Russia. Characteristic features include the fact that the fusoid-ellipsoid conidia are aseptate, and turn dark blue at maturity. Hercospora tiliae (also on Tilia cordata) represents a second genus in Lamproconiaceae, and is distinct in that conidia remain hyaline, and are aseptate. Our collections (on Tilia sp., Ukraine) differ in that the macroconidia are septate, and were not observed to become pigmented after 1 mo in culture.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence of CPC 34865 had highest similarity to Lamproconium desmazieri (as Melanconis desmazieri, strain MFLUCC 14-1047, GenBank KX430132.1; Identities = 530/577 (92 %), 19 gaps (3 %)), Sydowiella centaureii (voucher MFLU 16-2858, GenBank NR_155860.1; Identities = 500/552 (91 %), 19 gaps (3 %)), and Sydowiella fenestrans (strain CBS 125530, GenBank JF681956.1; Identities = 499/552 (90 %), 20 gaps (3 %)) – also see Fig. 39. The ITS sequence of CPC 34865 is identical to that of CPC 34862 (582/582 bases). Closest hits using the LSU sequence of CPC 34865 are Lamproconium desmazieri (as Melanconis desmazieri, strain AR3525, GenBank AF408372.1; Identities = 813/820 (99 %), no gaps), Hercospora tiliae (strain AR3526, GenBank AF408365.1; Identities = 790/820 (96 %), no gaps), and Diaporthe pustulata (strain AR3430, GenBank AF408357.1; Identities = 790/820 (96 %), no gaps) – also see Fig. 6, part 2b. The LSU sequence of CPC 34865 is identical to that of CPC 34862 (820/820 bases).
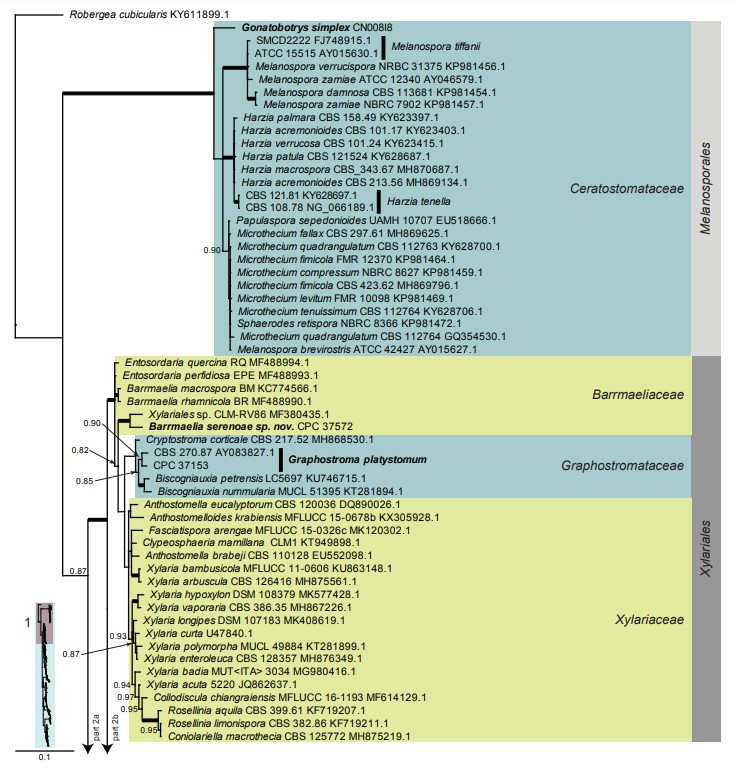
Fig. 6, parts 1–3. Consensus phylogram (50 % majority rule) resulting from a Bayesian analysis of the Sordariomycetes LSU sequence alignment. Bayesian posterior probabilities (PP) > 0.79 are shown at the nodes and the scale bar represents the expected changes per site. Thickened branches represent PP = 1. Families and orders are indicated with coloured blocks to the right of the tree. GenBank accession and/or culture collection numbers are indicated for all species. The tree was rooted to Robergea cubicularis (voucher G.M. 2013-05-09.1, GenBank KY611899.1) and the species treated in this study for which LSU sequence data were available are indicated in bold face. A miniature overview tree is also presented to facilitate navigation along the tree topology.
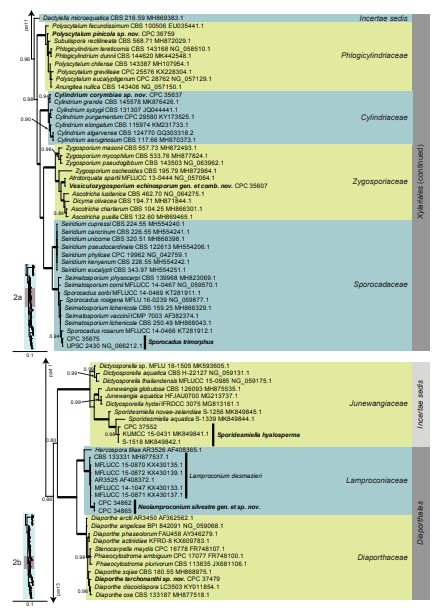
Fig. 6. (Continued).
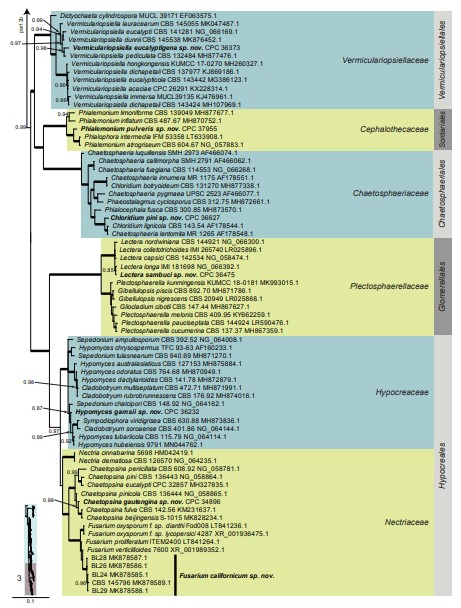
Fig. 6. (Continued).
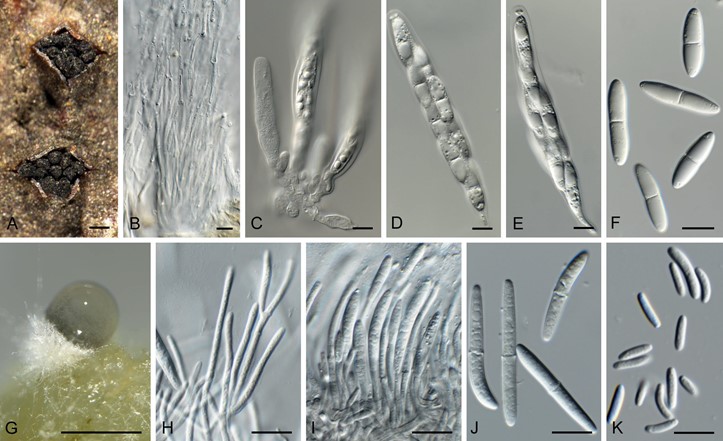
Fig. 38. Neolamproconium silvestre (CPC 34865). A. Stroma with ascomata. B. Paraphyses. C–E. Asci. F. Ascospores. G. Conidioma on OA. H, I. Conidiophores with conidiogenous cells. J. Macroconidia. K. Microconidia. Scale bars: A = 350 µm, G = 300 µm, all others = 10 µm.
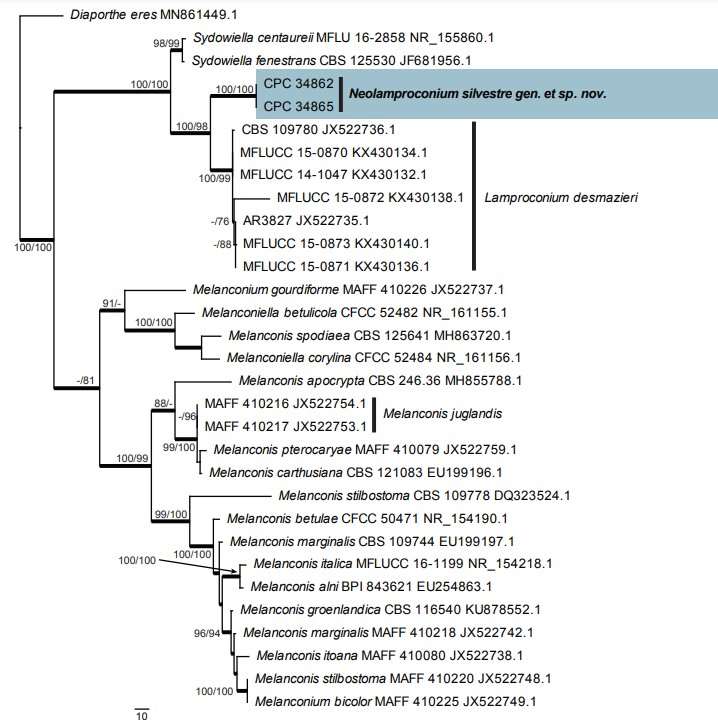
Fig. 39. The first of three equally most parsimonious trees obtained from a phylogenetic analysis of the “Lamproconium” ITS alignment (31 strains including the outgroup; 570 characters including alignment gaps analysed: 260 constant, 58 variable and parsimony-uninformative and 252 parsimony- informative). The tree was rooted to Diaporthe eres (strain UASWS2094, GenBank MN861449.1) and the scale bar indicates the number of changes. Parsimony bootstrap (PBS) and neighbour joining bootstrap support (NJBS) values higher than 74 % are shown at the nodes (PBS/NJBS) and the treated species is highlighted with a coloured box and bold text. Species names are indicated to the right of the tree, or before the culture collection and GenBank accession numbers. Branches present in the strict consensus tree are thickened. Tree statistics: TL = 686, CI = 0.724, RI = 0.917, RC = 0.664.
