Gymnopilus lepidotus Hesler, Mycologia Memoirs 3, 40 (1969)
Index Fungorum number: IF 314787; Facesoffungi number: FoF 14367; Fig. 1
Saprotrophic, solitary, growing on the rotting bark of a trunk of Cocos nucifera. Basidiomata small. Pileus 3–5 cm across, covex when young, plano-convex at maturity. Surface smooth, wet, yolk yellow (4B8) closer to the periphery, reddish-yellow (4A8) to orange (5A7) middle and center, covered reddish orange (7B8) erect scales, densely arranged at center and scattered towards margin. Margin rimose, hairy, edge slightly undulating at maturity, translucent sulcate, lamellae; deep yellow (4A8), deep orange (5A8), sub-decurrent, crowded with intermingled lamellulae of three lengths. Lamellulae smooth and thick, edge equal. Stipe 2.5–4.5 × 0.3–0.5 cm, surface reddish yellow (4A8) to dark orange (5B8) cylindric, slightly curved, flexible, pubscent, striated, flexible, tapering down, hollow, context: thin, 0.2–0.4 cm thick, deep yellow (4A8). Basidiospores 6.2–7.3 × 4.8–5.3 µm, ellipsoidal to amygdaliform, rounded to sub-acute apex, thin-walled, hyaline, warty. Basidia 18–23 × 6.2–7 µm, claviform, 2-4 spored, hyaline or sub-hyaline. Pleurocystidia 20–27 × 6.5–7.2 µm, utriform. Cheilocystidia lageniform with obtuse or sub-capitate apex, hyaline to sub-hyaline, hymenoophoral trama; thin-walled, clamp connections present.
Material examined – Sri Lanka, Southern Province, Matara District, growing on a rotting bark of a Cocos nucifera, 22 March 2021, A.N Ediriweera, FUOR0020AGS (HKAS123167); ibid., FUOR0076AGS (HKAS123152).
GenBank accession numbers – HKAS123152 – ITS: OQ607123; HKAS123167 – ITS: OQ607128.
Known habitat – Decomposing wood or occasionally terrestrial with buried or decomposed wood (Wright & Wright 2005, Lechner et al. 2006, Wright et al. 2008, Vasco-Palacios & Franco-Molano 2013, Grassi et al. 2016), rotting bark (this study).
Known distribution (based on molecular data) – Argentina (Lechner et al. 2006, Wright et al. 2008, Grassi et al. 2016), Colombia, Mexico (Vasco-Palacios & Franco-Molano 2013), USA (Wright & Wright 2005), Sri Lanka (this study).
Notes – This taxon has been recorded from Argentina in Misiones Province (Lechner et al. 2006, Wright et al. 2008, Grassi et al. 2016), Colombia and Mexico in the states of Jalisco and Veracruz (Guzmán-Dávalos 1996) and Florida, United States of America (Wright & Wright 2005). Our new strain is clustered with G. lepidotus isolates (XAL 30602 and XAL30374) with 70% maximum likelihood bootstrap support. Gymnopilus lepidotus formed a sister clade with G. purpuratus with 99% maximum likelihood bootstrap support and 0.99 Bayesian posterior probability. Fernando et al. (2015) reported G. lepidotus from Sri Lanka and studied its pharmacological properties. This study reports a new strain of G. lepidotus from Sri Lanka, which is phylogenetically closely related to the records from Mexico. The new strain differs by having a smaller pileus, a lower umbo at the center, absence of scales, presence of striates radiating up to the margin, and smaller basidiospores. Our new collection differs from the USA specimen in their basidiomata dimensions and the pileus size recorded as 4–8 mm. Guzmán-Dávalos et al. (2003) revised the taxa and found that the pileus diameter ranged from 6–18 (26) mm even in dried samples from Florida, while samples from Mexico extended up to 7 mm. Gymnopilus lepidotus reported from Paraguay showed very similar morphological characteristics to our new record.

Fig. 1 – Gymnopilus lepidotus (HKAS123167). a–c Mature basidiome. d–g Basidiospores. h, i Basidia. j, k Cheilocystidia. Scale bars: a–c = 0.5 cm, d–g = 2 µm, h–k = 5 µm.
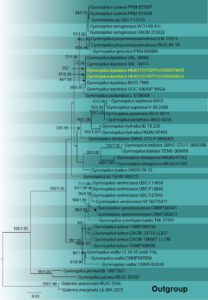
Fig. 2 – Phylogram generated from maximum likelihood analysis based on ITS sequence data of taxa in Gymnopilus. Forty-six strains are included in the phylogenetic analyses which comprised 1766 characters after alignment. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RaxML tree with a final likelihood value of -4446.715724. The matrix had 311 distinct alignment patterns, with 58.76% of undetermined characters or gaps. Evolutionary model applied for ITS is GTR+I+G. Bootstrap support values for ML equal to or greater than 65% and Bayesian posterior probabilities equal to or greater than 0.95 are indicated near the branches. The tree is rooted with Galerina autumnalis (IBUG 5246) and G. marginata (LE BIN 2272). Ex-type strains are in bold. The newly generated sequences are indicated in yellow.
