Gamszarea Z.F. Zhang & L. Cai, gen. nov.
Index Fungorum number: 556420, Facesoffungi num- ber: FoF 08439
Etymology: “Gamszarea” named in honour of Walter Gams and Rasoul Zare, for their contributions to the taxo- nomic study of Lecanicillium W. Gams & Zare.
Asexual morph: Conidiophores commonly arising from aerial hyphae, erect, hyaline. Conidiogenous cells discrete aculeate phialides, usually solitary or verticillate, some- times branched. Conidia adhering in more or less globose slimy heads and of two types, macroconidia first usually and then microconidia, aseptate. Macroconidia fusiform or falcate with more or less pointed ends; microconidia ellipsoidal, falcate, lunate or reniform. Crystals occasion- ally observed.
Sexual morph: only observed in Gamszarea wallacei on the pupal host. Perithecium hyaline, delicate, smooth, obclavate to naviculate. Asci 8-spored, with a prominent cap, narrowly cylindrical with an inflated vase. Ascospores hyaline, filiform, spirally twisted in the ascus, approximately the same length as the ascus, slender, indis- tinctly septate.
Type: Gamszarea wallacei (H.C. Evans) Z.F. Zhang & L. Cai
Notes: Lecanicillium was introduced by Gams and Zare (2001) to accommodate the taxa with aculeate phialides that cannot be classified in the genera such as Beauveria, Isaria Pers and Microhilum H.Y. Yip & A.C. Rath, with L. lecanii (Zimm.) Zare & W. Gams as the generic type (Sung et al. 2007a; Park et al. 2015; Huang et al. 2018). While, previous studies of Cordycipitaceae based on multi-locus phylogeny showed that Lecanicillium is polyphyly (Sung et al. 2007a; Sanjuan et al. 2014; Chiriví-Salomón et al. 2015; Kepler et al. 2017), and several species of Lecanicillium, including the type L. lecanii, were transferred to genus Akanthomyces Lebert (Kepler et al. 2017). Nevertheless, several distinctly separate clades remained (Figs. 25, 26). Three of our new species clustered with L. wallacei (H.C. Evans) H.C. Evans & Zare (teleomorph synonym: Torrubiella wallacei H.C. Evans), L. kalimantanense Kurihara & Sukarno, Verticillium indonesiacum Kurihara & Sukarno and several new Lecani- cillium species published recently in a single clade in Cordy- cipitaceae, which represented a new genus, herein named as Gamszarea (Figs. 25, 26). The most closely related genus to Gamszarea is Simplicillium Zare & W. Gams. Species of Simplicillium usually have discrete solitary phialides arising from prostrate hyphae and short-ellipsoidal to subglobose or obclavate conidia (Zare and Gams 2008). On contrary, phialides of Gamszarea are aculeate, solitary or verticillate and the dimorphic conidia are lunate, fusiform or falcate.
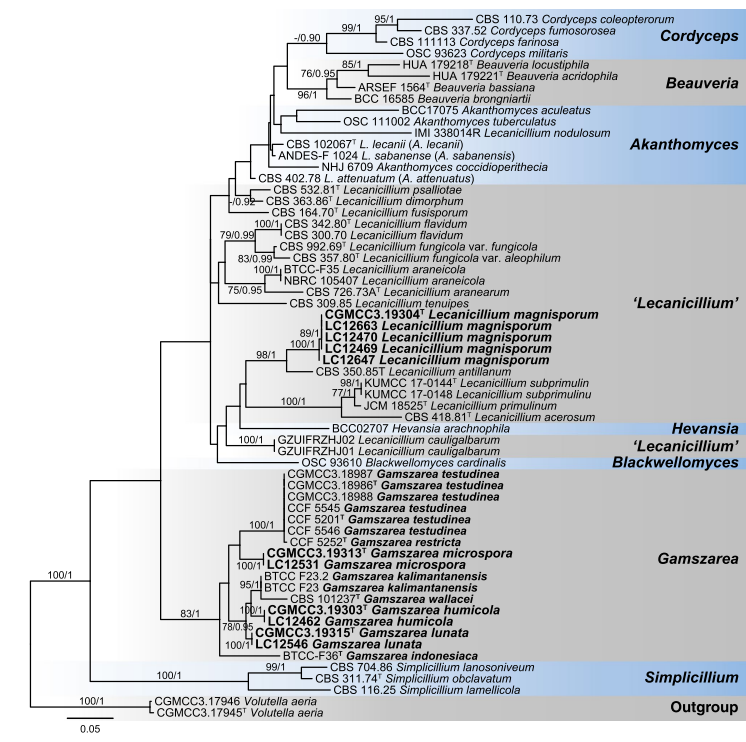
Fig. 25 Maximum likelihood (ML) tree of Gamszarea, Lecanicillium and allied genera in Cordycipitaceae based on ITS, LSU, SSU, EF1-α, RPB1 and RPB2 sequences. Seventy-six strains are used. The tree is rooted with Volutella aeria (CGMCC3.17945 and CGMCC3.17946). Tree topology of the ML analysis was similar to the BI. The Best scor- ing RAxML tree with a final likelihood value of − 41813.806368. The matrix had 2082 distinct alignment patterns, with 17.94 % of undeter- mined characters or gaps. Base frequencies estimated by jModelTest were as follows, A = 0.2338, C = 0.2762, G = 0.2605, T = 0.2295;substitution rates AC = 1.4660, AG = 3.7913, AT = 0.9486, CG = 0.9281, CT = 7.8283, GT = 1.0000; gamma shape = 0.5830. ML bootstrap values (≥ 70 %) and Bayesian posterior probability (≥ 90 %) are indicated along branches (ML/PP). Novel species are in bold font and “T” indicates type derived sequences
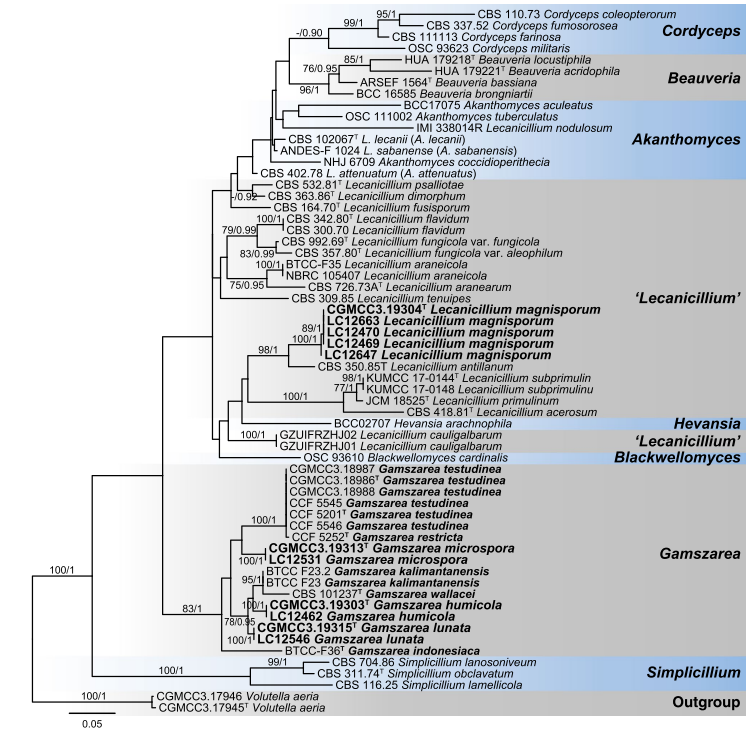
Fig. 26 Maximum likelihood (ML) tree of Gamszarea, Lecanicil- lium and allied genera in Cordycipitaceae based on ITS sequences. Sixty-two strains are used. The tree is rooted with Volutella aeria (CGMCC3.17945 and CGMCC3.17946). Tree topology of the ML analysis was similar to the BI. The Best scoring RAxML tree with a final likelihood value of − 5440.348928. The matrix had 347 dis- tinct alignment patterns, with 14.62 % of undetermined characters or gaps. Base frequencies estimated by jModelTest were as follows, A = 0.2220, C = 0.3155, G = 0.2645, T = 0.1980; substitution rates AC= 2.3755, AG = 2.4987, AT = 1.5316, CG = 0.9389, CT = 5.6398, GT = 1.0000; gamma shape = 0.5370. ML bootstrap values (≥ 70%) and Bayesian posterior probability (≥ 90 %) are indicated along branches (ML/PP). Novel species are in bold font and “T” indicates type derived sequences
Species
