Gamszarea wallacei (H.C. Evans) Z.F. Zhang & L. Cai, comb. nov.
MycoBank number: MB 556421; Index Fungorum number: IF 556421; Facesoffungi number: FoF 08440;
Basionym: Simplicillium wallacei H.C. Evans, Nova Hedwigia 73 (1–2): 43 (2001).
Synonym: Torrubiella wallacei H.C. Evans, Nova Hedwigia 73 (1–2): 46 (2001). Lecanicillium wallacei (H.C. Evans) H.C. Evans & Zare, Mycological Research 112 (7): 816 (2008).
Holotype: Indonesia, Sulawesi, Dumoga Bone forest, on lepidopteran larva, IMI 331549, ex-type living culture, CBS 101237.
Notes: This species was first described as Simplicillium wallacei by Gams and Zare (2001) based on morphological features, and then transferred to Lecanicillium based on ITS analyses (Zare and Gams 2008). While, in the cladogram of Zare and Gams (2008), Lecanicillium wallacei clustered in a distinct clade between Lecanicillium and Simplicillium, which was consistent with our multi-locus analyses (Figs. 25, 26). Therefore, a new combination is proposed here, as Gamszarea wallacei.
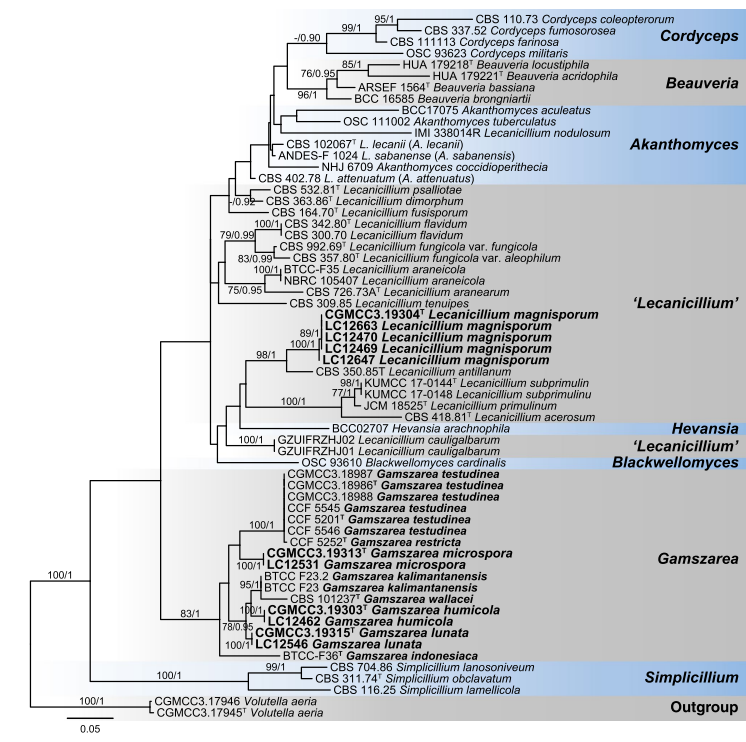
Fig. 25 Maximum likelihood (ML) tree of Gamszarea, Lecanicillium and allied genera in Cordycipitaceae based on ITS, LSU, SSU, EF1-α, RPB1 and RPB2 sequences. Seventy-six strains are used. The tree is rooted with Volutella aeria (CGMCC3.17945 and CGMCC3.17946). Tree topology of the ML analysis was similar to the BI. The Best scor- ing RAxML tree with a final likelihood value of − 41813.806368. The matrix had 2082 distinct alignment patterns, with 17.94 % of undeter- mined characters or gaps. Base frequencies estimated by jModelTest were as follows, A = 0.2338, C = 0.2762, G = 0.2605, T = 0.2295;substitution rates AC = 1.4660, AG = 3.7913, AT = 0.9486, CG = 0.9281, CT = 7.8283, GT = 1.0000; gamma shape = 0.5830. ML bootstrap values (≥ 70 %) and Bayesian posterior probability (≥ 90 %) are indicated along branches (ML/PP). Novel species are in bold font and “T” indicates type derived sequences
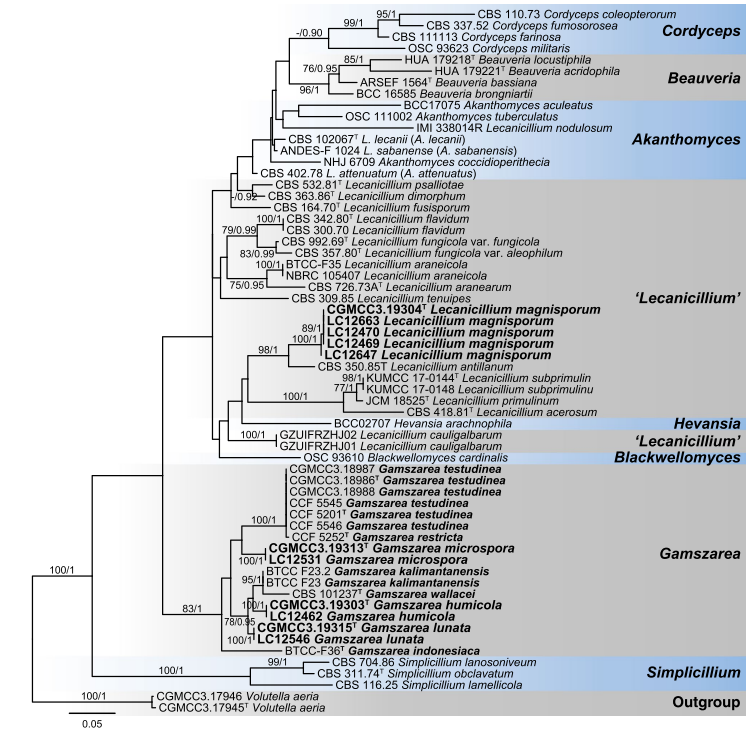
Fig. 26 Maximum likelihood (ML) tree of Gamszarea, Lecanicil- lium and allied genera in Cordycipitaceae based on ITS sequences. Sixty-two strains are used. The tree is rooted with Volutella aeria (CGMCC3.17945 and CGMCC3.17946). Tree topology of the ML analysis was similar to the BI. The Best scoring RAxML tree with a final likelihood value of − 5440.348928. The matrix had 347 dis- tinct alignment patterns, with 14.62 % of undetermined characters or gaps. Base frequencies estimated by jModelTest were as follows, A = 0.2220, C = 0.3155, G = 0.2645, T = 0.1980; substitution rates AC= 2.3755, AG = 2.4987, AT = 1.5316, CG = 0.9389, CT = 5.6398, GT = 1.0000; gamma shape = 0.5370. ML bootstrap values (≥ 70%) and Bayesian posterior probability (≥ 90 %) are indicated along branches (ML/PP). Novel species are in bold font and “T” indicates type derived sequences
