Salsuginea phoenicis S.N. Zhang, E.B.G. Jones, K.D. Hyde & J.K. Liu, in Jones et al., Botanica Marina 63(2), 158 (2019)
Index Fungorum number: IF 829547; Facesoffungi number: FoF 14242; Fig. 1
Saprobic on decaying petioles of Phoenix roebelenii. Sexual morph: Clypeus composed of host cells interspersed with darkened fungal tissue. Ascomata 400–520 μm high, 640–950 μm in diam. (x̄ = 489 × 802 µm, n = 10), solitary, scattered, immersed to erumpent, subglobose, black, carbonaceous, with a central ostiole, papillate, clypeate. Peridium thick, up to 50 μm wide, composed of brown cells of textura epidermoidea. Hamathecium comprising 1–2 μm wide, numerous, anastomosing, branched, septate, filiform, pseudoparaphyses, embedded in a gelatinous matrix. Asci 145–270 × 20–30 μm (x̄ = 229 × 27 µm, n = 20), 8-spored, bitunicate, fissitunicate, cylindric-clavate, pedicellate, with an apical apparatus consisting of a large distinctive ocular chamber. Ascospores 45–60 × 15–25 μm (x̅ = 55 × 20 μm, n = 20), uniseriate or overlapping uniseriate, hyaline when young, brown at maturity, broad fusiform or ellipsoidal, 2-celled and almost equally, strongly constricted at the central septum, with hyaline apical and basal germ pores, smooth-walled, lacking a mucilaginous sheath. Asexual morph: Undetermined.
Culture characteristics – Colonies on PDA attaining 3 cm diam. within 2 weeks at 25 °C under dark, velvety, circular, wavy margin, slightly raised, inner zone white and outer zone darker or pale brown, reverse brown, with a margin of translucent.
Material examined – China, Guangdong Province, Guangzhou City, Baiyun Mountain, near a small fresh water stream, on decaying, submerged petiole of Phoenix roebelenii, 16 August 2021, I.C. Senanayake, GZ14 (MHZU 22-0131), living cultures ZHKUCC 22-0226.
GenBank accession numbers – ZHKUCC 22-0226 – LSU: OQ586396, tef1-α: OQ590004; MHZU 22-0131 – LSU: OQ586396, tef1-α: OQ590004.
Known distribution (based on molecular data) – Thailand (Jones et al. 2020), China (this study).
Known hosts (based on molecular data) – on decaying petiole of Phoenix paludosa (Jones et al. 2020), on decaying, submerged petiole of P. roebelenii (this study).
Notes – The combined gene analysis of LSU, SSU, tef1-α, and rpb2 showed that our salsuginea-like collection (MHZU 22-0131) grouped with S. phoenicis with 96% maximum likelihood bootstrap support and 0.99 Bayesian posterior probability (Fig. 2). Our collection is morphologically very similar to the holotype of S. phoenicis (Jones et al. 2020). However, the holotype was collected from the marine environment, while ours’ was obtained from a freshwater habitat. Therefore, we identified this collection as S. phoenicis based on morphological and phylogenetic analyses (Figs. 1 and 2). This is the first report of S. phoenicis in China and the first collection after the holotype.

Fig. 1– Salsuginea phoenicis (MHZU 22-0131, a new geographical record). a, b Ascomata on the substrate. c–e Vertical cross-section of an ascoma. f Peridium. g Pseudoparaphyses. h–j Asci. k–n Ascospores. o Surface view of colony on PDA. p Reverse view of colony on PDA. Scale bars: d, e = 300 µm, f = 50 µm, g = 10 µm, h–j = 100 µm, k–n = 25 µm.
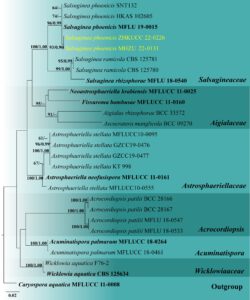
Fig. 2 – Phylogram generated from maximum likelihood analysis based on combined LSU, SSU, tef1-α, and rpb2 sequence data which comprised 3,669 characters (LSU = 899, SSU = 1020, tef1-α = 930, and rpb2 = 820). The best scoring RAxML tree with a final likelihood value of -8757.729573 is presented. The matrix had 422 distinct alignment patterns, with 7.32% of undetermined characters or gaps. Evolutionary model applied for all genes is GTR+I. Bootstrap support values for maximum likelihood equal to or greater than 60% and clade credibility values equal to or greater than 0.95 from Bayesian inference analysis are labelled at each node. Ex-type strains are in bold, while the new isolate is indicated in yellow. The tree is rooted to Caryospora aquatica (MFLUCC 11-0008).
