Magnibotryascoma mali Phukhams., Wanas. & K.D. Hyde, Fungal Diversity 87, 105 (2017)
Index Fungorum number: IF 553255; Facesoffungi number: FoF 003387; Fig. 1
Saprobic on dead branch of Osmanthus fragrans. Sexual morph: Undetermined. Asexual morph: Coelomycetous. Conidiomata 140–220 × 215–205 μm (x̄ = 185 × 250 μm, n = 20), solitary, semi-immersed or immersed in the substrate, globose to subglobose, dark brown to brown, with a short neck and a central ostiole. Peridium up to 11–26 μm wide, composed of dark brown to black, thick-walled cells of textura globosa, becoming thin-walled and hyaline towards the inner region. Paraphyses absent. Conidiophores subcylindrical to ampulliform, reduced to conidiogenous cells. Conidiogenous cells 2–7 × 2–4 μm (x̄ = 4 × 2 μm, n = 20), ampulliform to subcylindrical, hyaline, smooth-walled, arising from the inner layer of pycnidium wall. Conidia 3–5 × 2–3 μm (x̄ = 4 × 2 μm, n = 50), subglobose, oval, guttulate, hyaline when immature, golden brown at maturity, aseptate and smooth-walled.
Culture characteristics – Conidia germinating on PDA within 12 h. Colonies slow growing, reaching 30 mm diam. after 2 weeks at 20–25 °C, becoming ash-gray on the surface after one week, with the reverse side of the colonies pale gray to gray, and finally black after two weeks, dense, circular, slightly raised, surface smooth with crenate edge.
Material examined – China, Sichuan Province, Chengdu City, Chengdu Botanical Garden, 30°45’59”N, 104°7’19”E, on dead branch of Osmanthus fragrans, 28 March 2021, Na Wu, YW338, (GZAAS 22-2204), living culture GZCC 22-2202.
GenBank accession numbers – ITS: OP927796, LSU: OP926034, SSU: OP927796, tef1-α: OQ597716.
Known distribution (based on molecular data) – China (Hyde et al. 2017, this study), New Zealand (Crous et al. 2022b).
Known hosts (based on molecular data) – Malus halliana (Hyde et al. 2017), Metrosideros sp. (Crous et al. 2022b), Osmanthus fragrans (this study).
Notes – Our strain (GZCC 22-2202) clustered with the ex-type strain of Magnibotryascoma mali (MFLUCC 17-0933) based on the multi-gene phylogenic analyses (ITS, LSU, SSU, and tef1-α) (Fig. 2). Morphologically, our strain is similar to the M. mali (MFLU 17-0559) collected from decaying twigs of Malus halliana in China in having golden brown, oval, aseptate, smooth-walled conidia (Hyde et al. 2017). Based on morpho-molecular data analysis, we conclude that our new collection is a new host record of M. mali on Osmanthus fragrans in China.
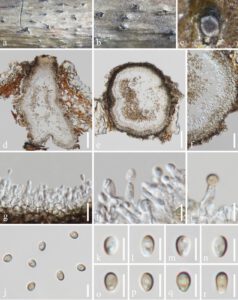
Fig. 1 – Magnibotryascoma mali (GZAAS 22-2204, new host record). a, b Conidiomata on host substrate. c–e Vertical section of a conidioma. f Section of the peridium. g–i Conidiogenous cells and developing conidia. j–r Conidia. Scale bars: d, e = 50 μm, f = 20 μm, g = 10 μm, h–r = 5 μm.

Fig. 2 – Phylogram generated from maximum likelihood analysis based on combined ITS, LSU, SSU and tef1-α sequence data for Teichosporaceae. Forty-three strains are included in the combined analyses which comprised 3290 characters (546 characters for ITS, 821 characters for LSU, 994 characters for SSU, 929 characters for tef1-α) after alignment. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RaxML tree with a final likelihood value of -10312.973943 is presented. Estimated base frequencies were as follows: A = 0.234738, C = 0.259620, G = 0.280304, T = 0.225338; substitution rates AC = 1.342287, AG = 2.282828, AT = 1.707394, CG = 1.224997, CT = 9.169275, GT = 1.000000; gamma distribution shape parameter α = 0.130805. The GTR+I+G evolutionary model was applied to all the genes in the dataset. Bootstrap support values for ML equal to or greater than 60% and Bayesian posterior probabilities equal to or greater than 0.95 are given near nodes, respectively. The tree was rooted with Torula chromolaee (MFLUCC 17-1504, MFLUCC 17-1514). Ex-type strains are in bold. The newly generated sequences are indicated in yellow.
