Robertozyma Q.M. Wang & F.Y. Bai gen. nov.
Index Fungorum number: MB 828824; Facesoffungi number: FoF
Etymology: the genus is named in honour of Dr. V. Robert for his contributions to the yeast taxonomy.
This genus is proposed for the branch represented by strain CGMCC 2.4451 which formed a separate clade. Member of the Cystobasidiales (Cystobasidiomycetes). The genus is mainly circumscribed by the phylogenetic analysis of the seven genes dataset, in which it occurred as a separate branch within the Cystobasidiales (Fig. 4).
Sexual reproduction not known. Colonies orange, butyrous. Budding cells present. Pseudohyphae and hyphae not produced. Ballistoconidia not formed.
Type species: Robertozyma ningxiaensis Q.M. Wang, F.Y. Bai & A.H. Li.
Note: – Robertozyma and its closely related genera, Beger- owomyces and Halobasidium, have a similar colony morphology, however, they can be distinguished by some physiological characters (Table S1.27). Robertozyma does not assimilate su- crose, melezitose, D-xylose and ethanol, whereas species of Begerowomyces and Halobasidium can use them. Beger- owomyces species assimilate erythritol and galactitol, whereas species of Robertozyma and Halobasidium do not assimilate these two carbon resources.
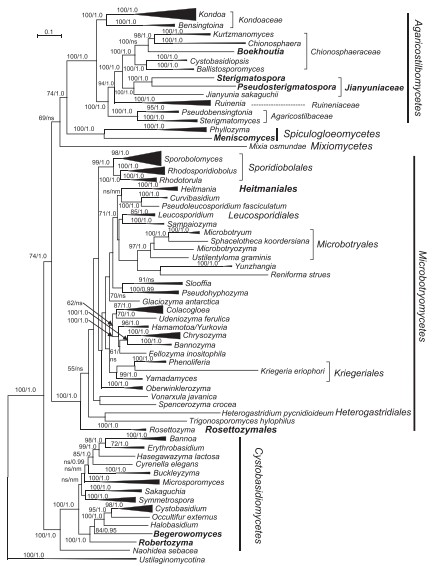
Fig. 4. Phylogenetic tree inferred using the combined sequences of RPB1, RPB2, TEF1, CYTB, SSU rDNA, LSU rDNA D1/D2 domains and 5.8S rDNA, depicting the phylogenetic positions of new taxa (in bold) within Pucciniomycotina. The tree backbone was constructed using maximum likelihood analysis. Bootstrap percentages of maximum likelihood analysis over 50 % from 1 000 bootstrap replicates and posterior probabilities of Bayesian inference above 0.9 are shown respectively from left to right on the deep and major branches. Bar = 0.2 substitutions per nucleotide position. Note: ns, not supported (BP < 50 % or PP < 0.9); nm, not monophyletic. The new taxa are in bold.
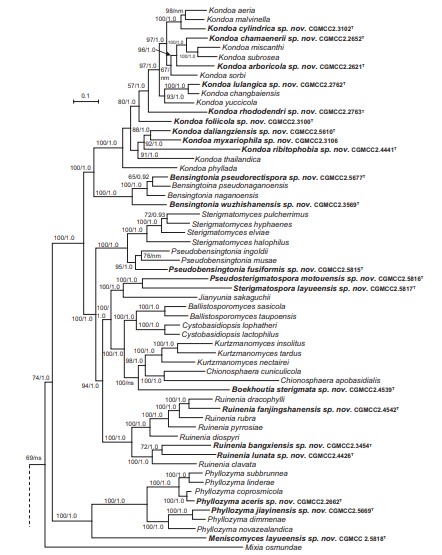
Fig. 4. (Continued).
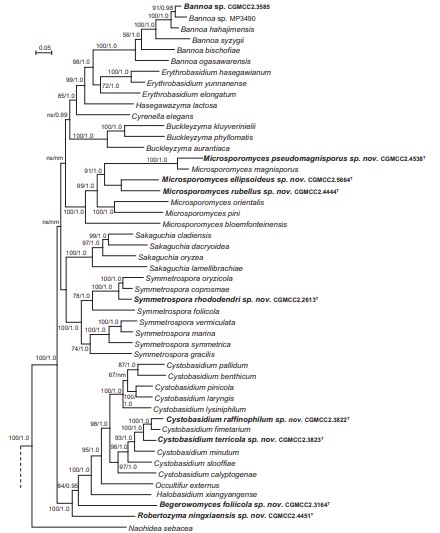
Fig. 4. (Continued).
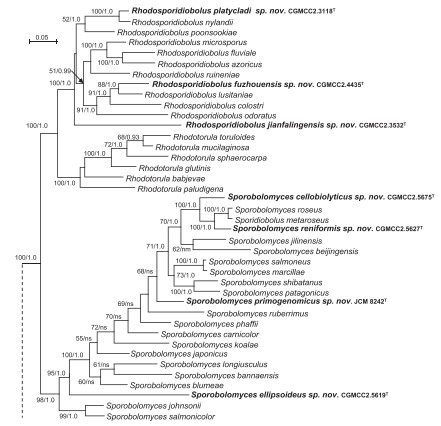
Fig. 4. (Continued).
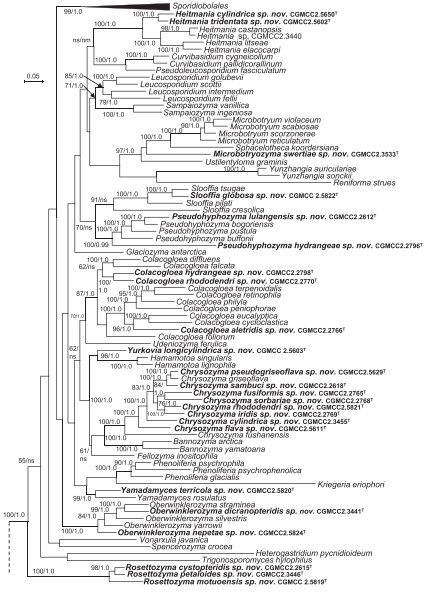
Fig. 4. (Continued).
Species
