Pseudoacrospermum goniomae Crous, sp. nov.
MycoBank number: MB 839304; Index Fungorum number: IF 839304; Facesoffungi number: FoF 12636; Fig. 61.
Etymology: Name refers to the host genus Gonioma from which it was isolated.
Mycelium consisting of septate, branched, smooth, pale brown, 2–2.5 µm diam hyphae. Conidiophores solitary, erect, flexuous, subcylindrical, multiseptate, unbranched with basal T-cell, arising from superficial mycelium, thick-walled, pale olivaceous, smooth, lacking rhizoids, 100–180 × 2.5–3.5 µm, terminating in a slightly swollen conidiogenous cell. Conidiogenous cells integrated, terminal, subclavate, 15–20 × 4–6 µm, with numerous sympodial loci aggregated in a rachis, darkened, thickened, but not refractive, pimple-like, protruding (0.5 µm), 1–2 µm diam. Conidia solitary, smooth, pale olivaceous to granular, subclavate, curved, apex obtuse, tapering to truncate hilum, 2 µm diam, darkened, thickened, and refractive, somewhat thick-walled, 3(–6)-septate, (17–)21–23(–25) × (5–)6 µm.
Culture characteristics: Colonies flat, spreading, surface folded, with moderate aerial mycelium and smooth, even margin, reaching 5 mm diam after 2 wk at 25 °C. On MEA surface buff to honey, reverse isabelline; on PDA surface straw in centre, outer margin buff to cinnamon, reverse cinnamon; on OA surface cinnamon.
Typus: South Africa, Western Cape Province, Knysna, on leaves of Gonioma kamassi (Apocynaceae), 22 Nov. 2018, F. Roets, HPC 2733 (holotype CBS H-24393, culture ex-type CPC 37030 = CBS 146732).
Notes: Pseudoacrospermum is phylogenetically closely related to Acrospermum, which is known only by a sexual morph. Based on its phylogeny, Acrospermum has been shown to be closely related to such as Gonatophragmium, Radulidium or Pseudovirgaria (Hudson et al. 2019), that are presently known by their asexual morphs. Pseudoacrospermum is distinguished based on its multiseptate conidia. There is no evidence to suggest that Pseudoacrospermum can be linked to Acrospermum. A phylogenetic tree is presented as Fig. 62.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Acrospermum maxonii (as Acrospermum sp. VD- 2019c; voucher 2711, GenBank MK562001.1; Identities = 344/421 (82 %), 23 gaps (5 %)), Radulidium epichloes (strain CBS 115704, GenBank MH862990.1; Identities = 217/236 (92 %), five gaps (2 %)), and Acrospermum leucocephalum (as Acrospermum sp. VD-2019b; voucher 764, GenBank MK562008.1; Identities = 334/413 (81 %), 22 gaps (5 %)). Closest hits using the LSU sequence are Acrospermum gorditum (as Acrospermum sp. VD-2019a; voucher 1774, GenBank MK561981.1; Identities = 803/867 (93 %), seven gaps (0 %)), Acrospermum maxonii (as Acrospermum sp. VD-2019c; voucher 3114, GenBank MK561987.1; Identities = 804/870 (92 %), 11 gaps (1 %)), and Gonatophragmium triuniae (strain CBS 138901, GenBank NG_058117.1; Identities = 764/827 (92 %), nine gaps (1 %)) – also see Fig. 1. The best hit using the rpb2 sequence had highest similarity to Pseudovirgaria hyperparasitica (strain CBS 121739, GenBank XM_033741841.1; Identities = 573/769 (75 %), ten gaps (1 %)). Only very distant hits with taxa in Dothideomycetes were obtained using the tef1 (first and second part) sequences. However, both these sequences were manually compared with blast2 searches against Acrospermum tef1 sequences labelled as unverified in the GenBank nucleotide database: Closest hits using the tef1 (first part) sequence had highest similarity to Acrospermum maxonii (strain 75017, GenBank MK604769.1; Identities = 198/234 (85 %), seven gaps (2 %)), Acrospermum gorditum (strain 1774, GenBank MK604778.1; Identities = 196/232 (84 %), two gaps (0 %)), and Acrospermum leucocephalum (strain 3342, GenBank MK604780.1; Identities = 213/256 (83 %), nine gaps (3 %)). Closest hits using the tef1 (second part) sequence had highest similarity to Acrospermum maxonii (strain 44, GenBank MK604767.1; Identities = 426/473 (90 %), no gaps), and Acrospermum leucocephalum (strain 764, GenBank MK604781.1; Identities = 412/474 (87 %), one gap (0%)).
Authors: P.W. Crous, J.Z. Groenewald & F. Roets
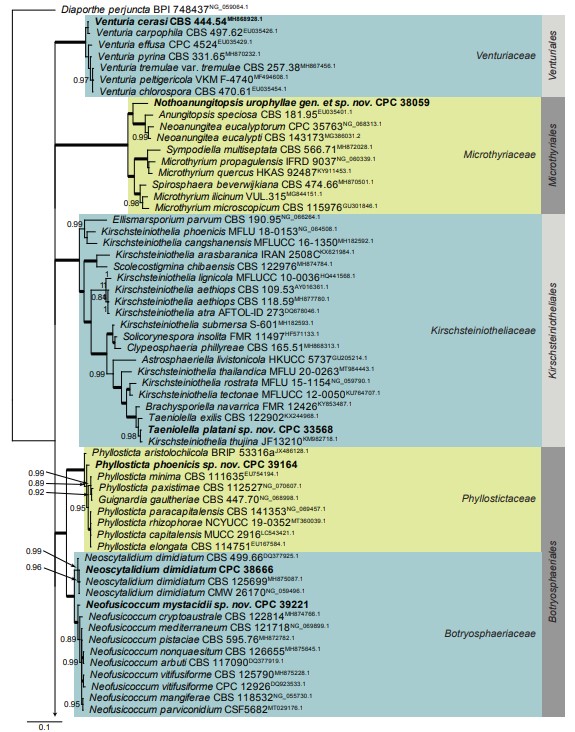
Fig. 1, parts 1–5. Consensus phylogram (50 % majority rule) resulting from a Bayesian analysis of the Dothideomycetes LSU nucleotide alignment. Bayesian posterior probabilities (PP) > 0.79 are shown at the nodes and the scale bar represents the expected changes per site. Thickened branches represent PP = 1. The branch leading to Superstratomycetales was halved to facilitate layout. Families and orders are indicated with coloured blocks to the right of the tree. GenBank accession (superscript) and / or culture collection / voucher numbers are indicated for all species. The tree was rooted to Diaporthe perjuncta (voucher BPI 748437, GenBank NG_059064.1) and the species treated in this study for which LSU sequence data were available are indicated in bold face.

Fig. 1. (Continued).
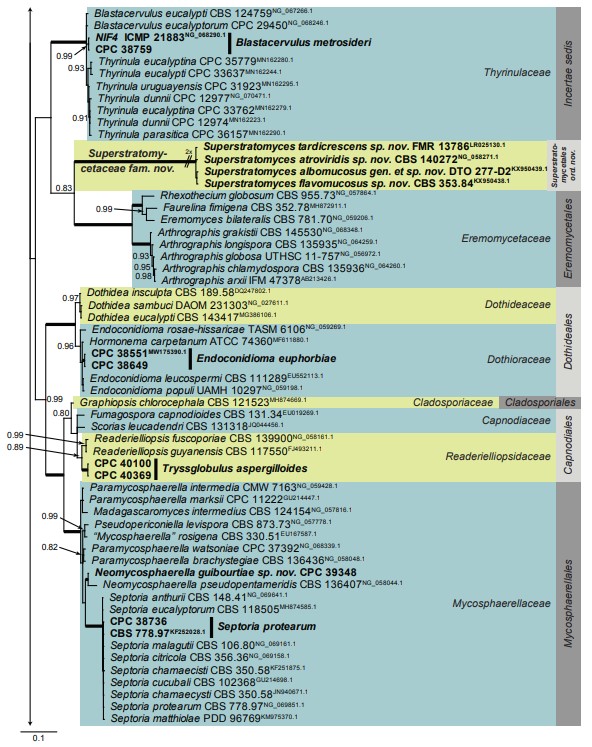
Fig. 1. (Continued).
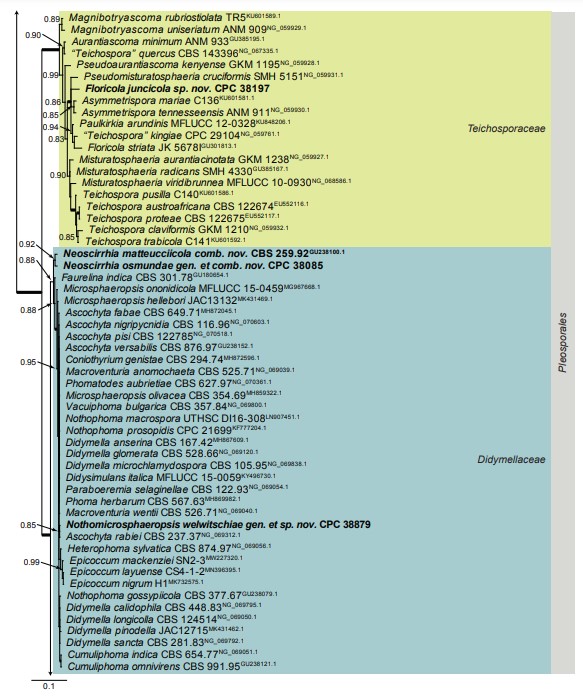
Fig. 1. (Continued).
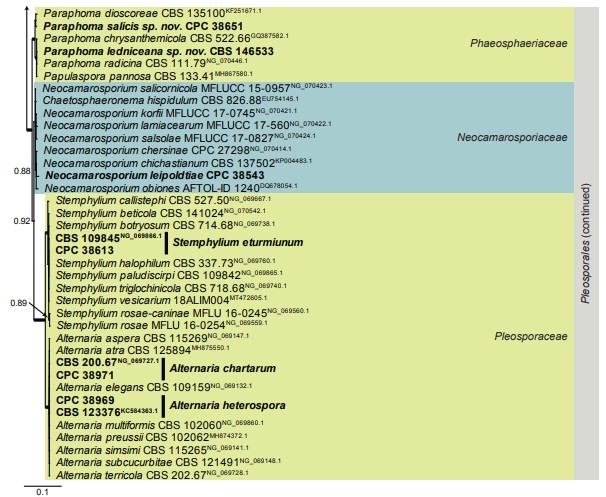
Fig. 1. (Continued).
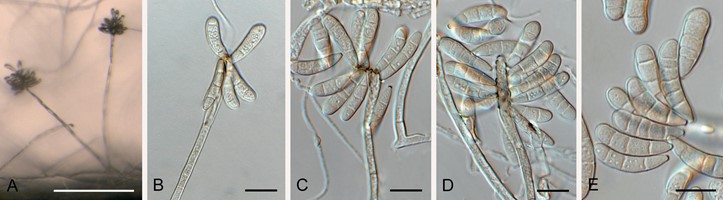
Fig. 61. Pseudoacrospermum goniomae (CPC 37030). A. Conidiophores on SNA. B–D. Conidiogenous cells giving rise to conidia. E. Conidia. Scale bars: A = 180 µm, all others = 10 µm.
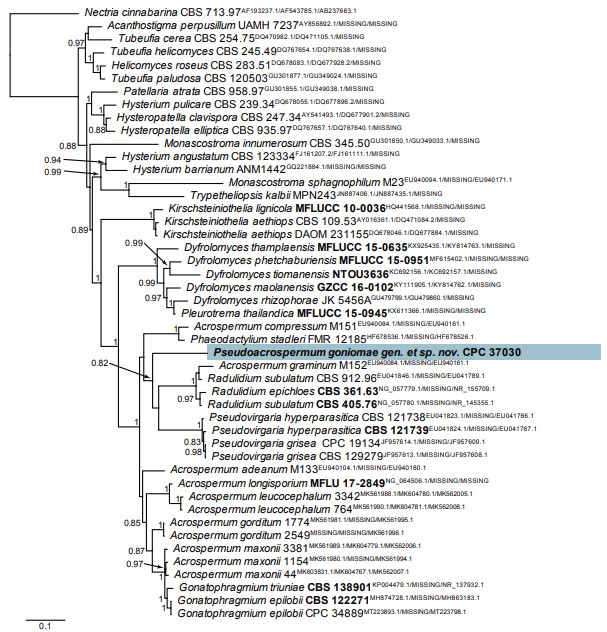
Fig. 62. Consensus phylogram (50 % majority rule) resulting from a Bayesian analysis of the Acrospermum and related genera multigene (LSU / tef1 / ITS) nucleotide alignment. The alignment is derived from Hudson et al. (2019). Bayesian posterior probabilities (> 0.79) are shown at the nodes and the scale bar represents the expected changes per site. GenBank accession (superscript) and / or culture collection / voucher numbers (in bold face when having a type status) are indicated for all species. The tree was rooted to Nectria cinnabarina (culture CBS 713.97) and the novel species treated in this study is indicated in a coloured block and bold face. Alignment statistics: 47 strains including the outgroup; 595 / 418 / 284 unique site patterns, respectively. Tree statistics: 184 502 sampled trees from 1 230 000 generations.
