Nothomicrosphaeropsis welwitschiae Crous, sp. nov.
MycoBank number: MB 839299; Index Fungorum number: IF 839299; Facesoffungi number: FoF; Fig. 43.
Etymology: Named after the host genus it was collected from, Welwitschia.
Conidiomata pycnidial, solitary, erumpent, brown, 200–250 µm diam, papillate with central ostiole; wall of 3–6 layers of brown textura angularis. Conidiophores reduced to conidiogenous cells lining the inner cavity, ampulliform to doliiform, hyaline, smooth, 4–6 × 3–4 µm. Conidia solitary, hyaline, smooth, aseptate, granular, subcylindrical, apex obtuse, base bluntly rounded, straight to slightly curved, becoming pale brown with age, (4–)5–6(–7) × 2–2.5 µm.
Culture characteristics: Colonies flat, spreading, with moderate aerial mycelium and smooth, even margin, covering dish after 2 wk at 25 °C. On MEA, PDA and OA surface and reverse olivaceous grey.
Typus: Namibia, Hope Mine, east of Gobabeb – Namib Research Institute, on dead leaves of Welwitschia mirabilis (Welwitschiaceae), 19 Nov. 2019, P.W. Crous, HPC 3126 (holotype CBS H-24448, culture ex-type CPC 38879 = CBS 146829).
Notes: Based on ITS/LSU DNA sequence data, the present collection was initially identified as Macroventuria wentii, which is known from plant litter in the Nevada Death Valley, USA (van der Aa 1971). However, by incorporating more informative genes such as rpb2 and tub2 in the analysis (see phylogenetic tree, Fig. 44), the present isolate proved allied to Paramicrosphaeropsis, Neomicrosphaeropsis and Microsphaeropsis, a generic complex in Didymellaceae (Hou et al. 2020a). Genera in this complex are morphologically comparable in initially producing hyaline conidia that turn pale brown at maturity.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to “Aff. Phoma sp.” (strain mk175, GenBank KT150686.1; Identities = 530/530 (100 %), no gaps), Macroventuria wentii (strain CBS 526.71, GenBank MH860250.1; Identities = 538/540 (99 %), no gaps), and Leptosphaerulina australis (strain CBS 234.58, GenBank MH857766.1; Identities = 538/542 (99 %), one gap (0 %)). Closest hits using the LSU sequence are Ascochyta rabiei (strain CBS 237.37, GenBank NG_069312.1; Identities = 858/858 (100 %), no gaps), Coniothyrium genistae (strain CBS 294.74, GenBank MH872596.1; Identities = 860/861 (99 %), no gaps), Ascochyta fabae (strain CBS 649.71, GenBank MH872045.1; Identities = 860/861 (99 %), no gaps), and Macroventuria wentii (strain CBS 526.71, GenBank NG_069040.1; Identities = 856/858 (99 %), no gaps) – also see Fig. 1. Closest hits using the rpb2 sequence had highest similarity to Neomicrosphaeropsis italica (strain IT_24211, GenBank KU714604.1; Identities = 693/744 (93 %), no gaps), Neomicrosphaeropsis juglandis (strain MFLUCC 18-0795, GenBank MN593307.1; Identities = 693/744 (93%), no gaps), and Neomicrosphaeropsis cytisi (strain MFLUCC 13-0396, GenBank KX572355.1; Identities = 682/742 (92 %), no gaps). A direct blast2 search against Macroventuria rpb2 sequences on GenBank had highest similarity to Macroventuria anomochaeta (strain CBS 525.71, GenBank GU456346.1; Identities = 663/741 (89 %), no gaps), Macroventuria terrestris (strain RMF CA 12, GenBank MT018194.1; Identities = 531/596 (89 %), no gaps), and Macroventuria wentii (strain CBS 527.71C, GenBank MN983625.1; Identities = 530/596 (89 %), no gaps). Closest hits using the tub2 sequence had highest similarity to Paramicrosphaeropsis ellipsoidea (strain CBS 197.97, GenBank MT005680.1; Identities = 314/333 (94 %), no gaps), Epicoccum sorghinum (strain FDY-5, GenBank MT799852.1; Identities= 336/364 (92 %), no gaps), Epicoccum latusicollum (strain HH12, GenBank MN623288.1; Identities = 337/365 (92 %), two gaps (0 %)), and Macroventuria wentii (strain CBS 526.71, GenBank GU237546.1; Identities = 302/333 (91 %), no gaps). A direct blast2 search against Macroventuria tub2 sequences on GenBank had highest similarity to Macroventuria wentii (strain CBS 877.70, GenBank MN984003.1; Identities = 303/333 (91 %), no gaps), Macroventuria terrestris (strain CBS 127771, GenBank MT005653.1; Identities = 300/331 (91 %), one gap (0 %)), and Macroventuria anomochaeta (strain CBS 525.71, GenBank GU237545.1; Identities = 300/333 (90 %), no gaps).
Authors: P.W. Crous, D.A. Cowan, G. Maggs-Kölling, E. Marais, N. Yilmaz & M.J. Wingfield
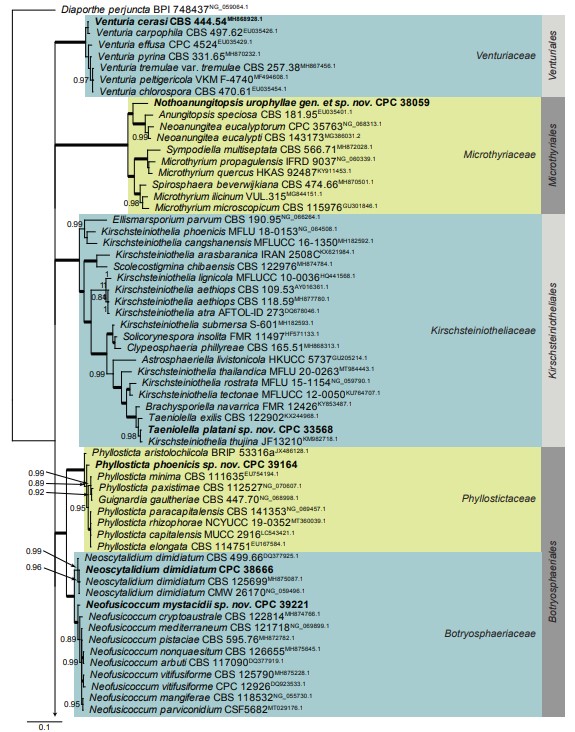
Fig. 1, parts 1–5. Consensus phylogram (50 % majority rule) resulting from a Bayesian analysis of the Dothideomycetes LSU nucleotide alignment. Bayesian posterior probabilities (PP) > 0.79 are shown at the nodes and the scale bar represents the expected changes per site. Thickened branches represent PP = 1. The branch leading to Superstratomycetales was halved to facilitate layout. Families and orders are indicated with coloured blocks to the right of the tree. GenBank accession (superscript) and / or culture collection / voucher numbers are indicated for all species. The tree was rooted to Diaporthe perjuncta (voucher BPI 748437, GenBank NG_059064.1) and the species treated in this study for which LSU sequence data were available are indicated in bold face.

Fig. 1. (Continued).
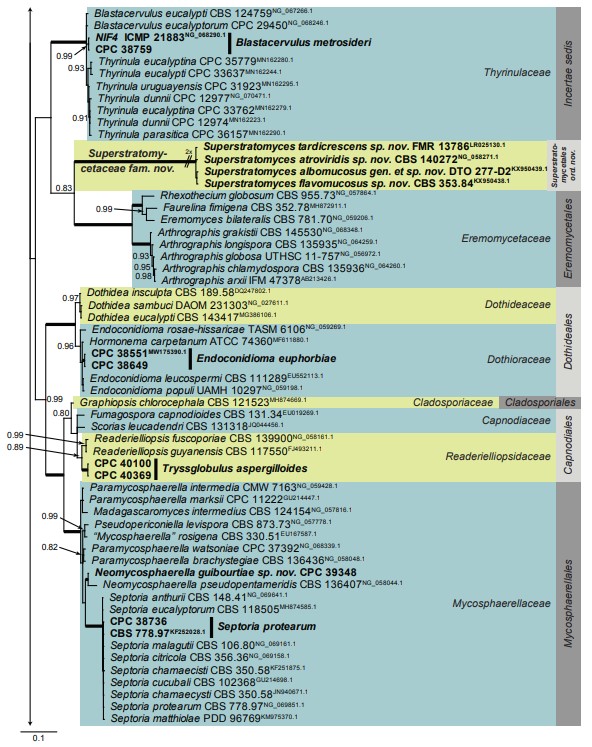
Fig. 1. (Continued).
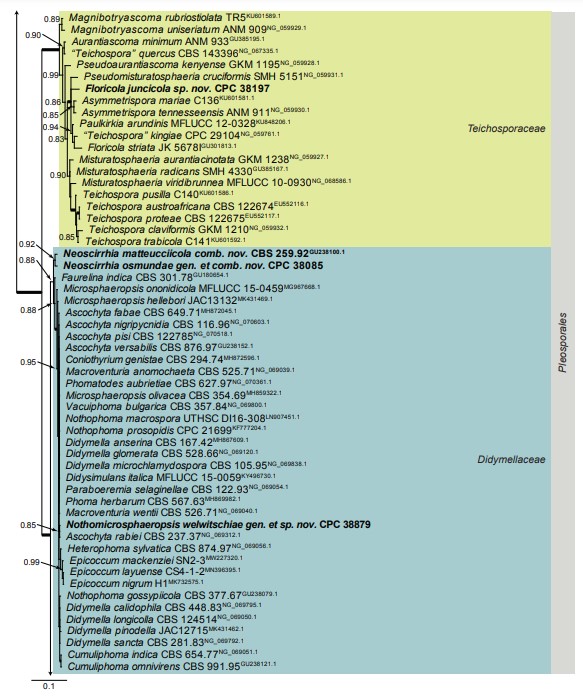
Fig. 1. (Continued).
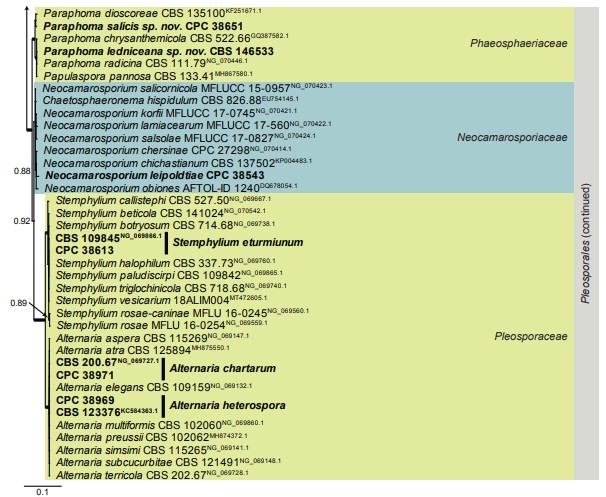
Fig. 1. (Continued).
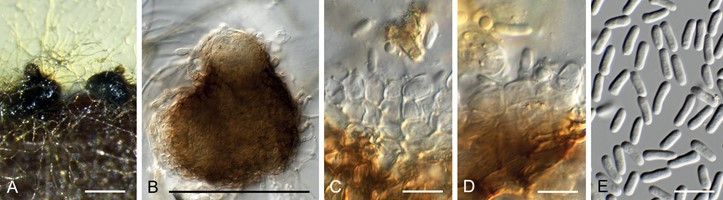
Fig. 43. Nothomicrosphaeropsis welwitschiae (CPC 38879). A. Conidiomata on PNA. B. Pycnidial conidioma. C–D. Conidiogenous cells. E. Conidia. Scale bars: A–B = 250 µm, all others = 10 µm.
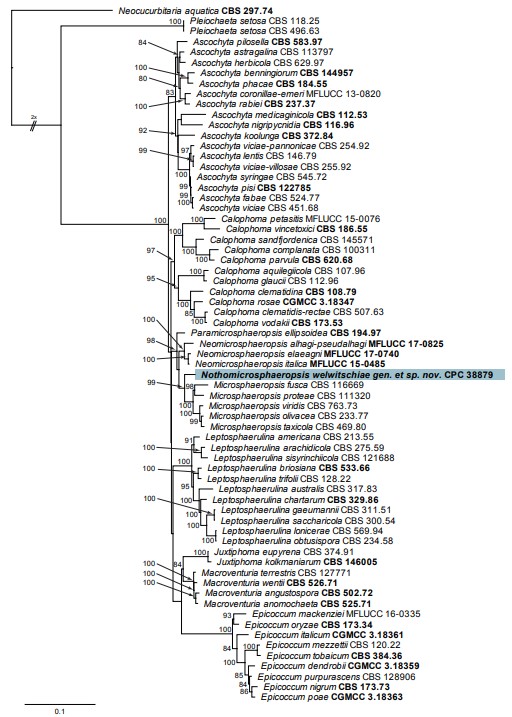
Fig. 44. Consensus phylogram (50 % majority rule) obtained from the maximum likelihood analysis with IQ-TREE showing the phylogenetic position of Nothomicrosphaeropsis welwitschiae gen. et sp. nov. based on the multigene (ITS / LSU / rpb2 / tub2) nucleotide alignment. The most basal branch was halved to facilitate layout. Bootstrap support values (> 79 %) from 5 000 ultrafast bootstrap replicates are shown at the nodes. The alignment is derived from the combined alignment of Hou et al. (2020a) and GenBank accession numbers can be obtained from the same reference. Culture collection / voucher numbers (in bold face when having a type status) are indicated for all species. The tree was rooted to Neocucurbitaria aquatica (culture CBS 297.74) and the species treated in this study is highlighted with a coloured block and bold face. Alignment statistics: 67 strains including the outgroup; 2 382 characters including alignment gaps analysed: 625 distinct patterns, 505 parsimony-informative, 94 singleton sites, 1 783 constant sites. The best models identified in IQ-TREE were: TIM2e+I+G4 (ITS), TNe+I+G4 (LSU), TNe+I+G4 (rpb2), TIM2e+I+G4 (tub2).
