Paramonodictys yunnanensis E.F. Yang & S. Tibpromma, sp. nov
MycoBank number: MB; Index Fungorum number: IF; Facesoffungi number: FoF 10591;
Holotype: KUMCC 21-0337
Saprobic on dead branch of Mangifera indica. Sexual morph: Undetermined. Asexual Morph: Colonies on substrate effuse, superfcial, erect, solitary, obovoid to oblong, visible brownish to dark brown dots scattered on the host surface, easily separating when disturbed. Mycelium fully immersed. Conidiophores reduced to Conidiogenous cells 8–12 (x̅ = 10 μm, n = 20) in high, 9–12 (x̅ = 10.6 μm, n = 20) in diam., brownish to reddish-brown, cylindrical. Conidia 47–70 × 35–47 μm (x̅ = 58 × 41 μm, n = 20), obovoid to subglobose, muriform, thick-walled, irregularly multiseptate, brown to olivaceous brown when mature, not in chains.
Culture characteristics: Conidia germinated within 18–20 hours on PDA, Colonies rapid-growing on PDA reaching about 10–15 mm diam. after 2 week at room temperature in natural light, colony circular, umbonate, fluffy, gray to brown from above, dark-brown from below, erose to dentate, without pigments produced in PDA.
Material examined: CHINA, Yunnan Province, Honghe Menglong village on dead branch of Mangifera indica, (102 ° 50’11”E, 23°41’01’’N, 500.6 m), 22 December. 2020, E.F. Yang, HHE001, Herb. KUN-HKAS 122189, culture holotype KUMCC 21-0337 culture ex-type KUMCC KUMCC 21-0347.
Note: The morphological characteristics of Paramonodictys yunnanensis sp. nov. tally with described for Paramonodictys genus in having brown to olivaceous-brown conidia attachment on superficial of substrate, solitary, obovoid to oblong, high similarity in size, however the conidiogenous cells of our novel P. yunnanensis is shorter than P solitarius, the colonies on PDA has distinct difference with P. solitarius MFLUCC 17-2353 ( brown vs. olivaceous brown; erose margin vs. entire edge). In the NCBI blastn of ITS sequence, our strain P. yunnanensis KUMCC 21-0337 highly overlapped with P. solitarius (strains: MFLUCC 17-2353 and GZCC 20-0007) at the 98.97% and 98.74% similarity. Moreover, based on SSU, LSU, ITS and tef1-a dataset, the multi-gene phylogeny generated herein indicates that P. yunnanensis close to P. solitarius branches with high bootstrap sport value in ML and BI phylogenetic trees, but they were separate well. Therefore, our isolation as new species be established in this study.
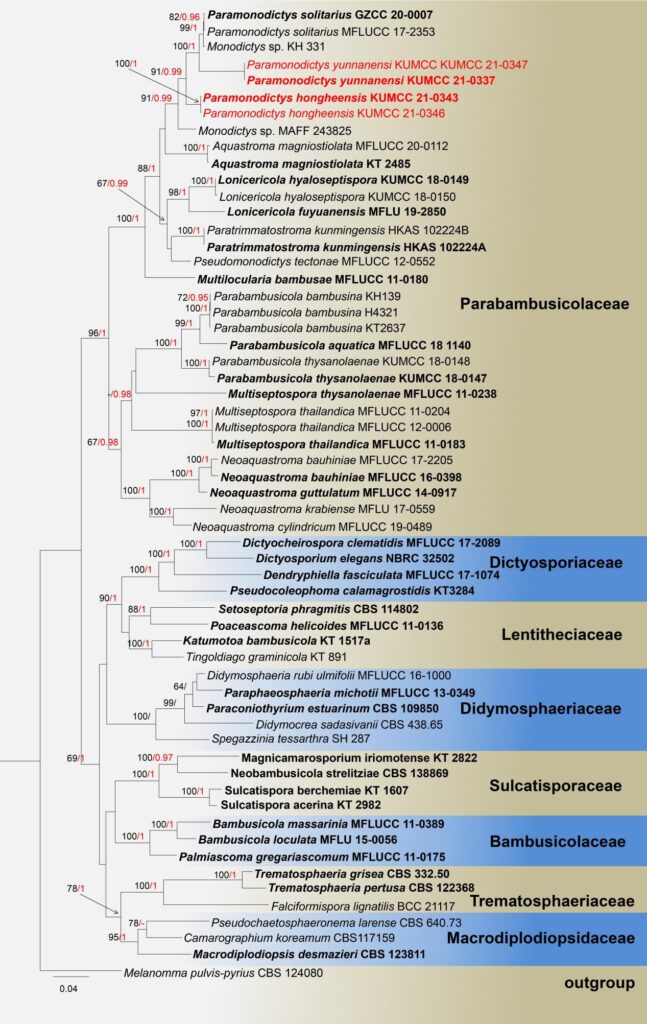
Fig.X. Phylogram generated from Maximum Likelihood analysis based on a combined SSU, LSU, ITS, and tef1-a sequence datasets, 59 strains are included in the combined gene analyses 3540 total character including gaps (LSU:1-875 bp, SSU:880-1903 bp, tef1-a: 1908-2837 bp, ITS: 2842-3540 bp), Maximum likelihood bootstrap support values greater than 60% are shown in above branches, Tree topology of the ML analysis was similar to the BI. The matrix had distinct alignment patterns, with the final ML optimization likelihood value of -27170.920818 (ln). All free model parameters were estimated by RAxML model, with 1459 distinct alignment patterns and 21.90% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.235389, C = 0.254603, G = 0.273057, T = 0.236951, with substitution rates AC = 1.064440, AG = 2.450499, AT = 1.326228, CG = 1.102216, CT = 5.326081, GT = 1.000000. The gamma distribution shape parameter alpha = 0.516907 and the Tree-Length = 3.201655. The final average standard deviation of split frequencies at the end of total MCMC generations calculated as 0.009940 in BI analysis. while Bayesian posterior probabilities greater than 0.95 are in bold. Tree topology of the ML analysis was similar to the BI. the type species bold and new isolates in red bold.

Fig. X. Paramonodictys yunnanensi (KUN-HKAS 122189, holotype) a, b. Close-up of colonies on substrate; c, d. Conidia attached on host; e. Single germinated conidium; f, g. Colony from above and below; h–k conidia connect with conidiogenous cells; l–m. immature and mature conidia.— Scale bars: c =100 μm; d, e, h–o = 50 μm.
