Nothoseptoria caraganae (Henn.) Crous & Bulgakov, comb. nov.
MycoBank number: MB 835095; Index Fungorum number: IF 835095; Facesoffungi number: FoF; Fig. 43.
Basionym: Septoria caraganae Henn., Z. PflKrankh. 12: 15. 1902.
Synonym: Mycosphaerella jaczewskii Potebnia, Annls mycol. 8(1): 50. 1910.
Leaf spots pale brown, confined by leaf veins, angular, leading to leaf blight. Conidiomata pycnidial, hypophyllous on leaves, oozing a creamy conidial cirrhus. Conidiomata brown, pycnidial, 250–300 µm diam, opening via central ostiole; wall of 3–6 layers of brown textura angularis. Conidiophores reduced to conidiogenous cells lining the inner cavity, hyaline, smooth, ampulliform to subcylindrical, 6–10 × 3–5 µm, proliferating percurrently at apex. Conidia solitary, straight to slightly curved, subcylindrical, apex subobtuse, base truncate with minute marginal frill, 1(–3)-septate, prominently guttulate, (35–)41–50(–60) × (3.5–)4(–5.5) µm, in culture (32–)35–40(–45) × (3–)4 µm.
Culture characteristics: Colonies erumpent, spreading, with sparse aerial mycelium and smooth, lobate margin, reaching 3 mm diam after 2 wk at 25 ºC. On MEA, PDA and OA surface and reverse olivaceous grey.
Materials examined: Russia, Rostov region, Shakhty city district, Hospital park, on live leaves of Caragana arborescens (Leguminosae), 21 Sep. 2018, T.S. Bulgakov, HPC 2631 = Myc-01 = CBS H-24229, culture CPC 36563 = CBS 145993; Rostov region, Shakhty city district, street shrubs, on live leaves of Caragana arborescens, 21 Sep. 2018, T.S. Bulgakov, HPC 2632 = Myc-02, culture CPC 36565.
Notes: Septoria caraganae was described from leaves of Caragana arborescens collected in Germany, with conidia being 1–3-septate, 30–50 × 3–4 µm (von Hennings 1902). The link to the sexual morph, Mycosphaerella jaczewskii, requires confirmation in culture.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence of CPC 36563 had highest similarity to Sonderhenia radiata (strain CBS 145600, GenBank MN162023.1; Identities = 435/490 (89 %), 10 gaps (2 %)), Sonderhenia eucalyptorum (strain CPC 32601, GenBank MN162022.1; Identities = 435/490 (89 %), 10 gaps (2%)), and Sonderhenia eucalypticola (strain CPC 31596, GenBank MN162018.1; Identities = 434/490 (89 %), 10 gaps (2 %)). The ITS sequences of CPC 36563 and 36565 are identical (485/485 bases). Closest hits using the LSU sequence of CPC 36563 are Sonderhenia eucalyptorum (strain CPC 17677, GenBank MN162214.1; Identities = 805/836 (96 %), 9 gaps (1 %)), Sonderhenia eucalypticola (as Mycosphaerella walkeri, strain CMW 20333, GenBank DQ267574.1; Identities = 803/834 (96 %), 5 gaps (0 %)), and Fusoidiella depressa (as Passalora depressa, strain CPC 14915, GenBank KF251813.1; Identities = 790/823 (96 %), 8 gaps (0 %)) – also Fig. 3, part 2 and Fig. 8, part 1. The LSU sequences of CPC 36563 and 36565 are identical (834/834 bases). Closest hits using the rpb2 sequence of CPC 36563 had highest similarity to Septoria protearum (strain CBS 778.97, GenBank KT216543.1; Identities = 750/924 (81 %), 12 gaps (1%)), Septoria bupleuricola (strain TCM-5, GenBank KY798146.1; Identities = 730/921 (79 %), 6 gaps (0 %)), and Mycovellosiella cajani (strain CBS 113999, GenBank MF951528.1; Identities = 732/928 (79 %), 18 gaps (1 %)). The rpb2 sequences of CPC 36563 and 36565 are identical (892/892 bases). No significant hits were obtained when the tub2 sequence of CPC 36563 was used in blastn and megablast searches. The tub2 sequences of CPC 36563 and 36565 are identical (436/436 bases).
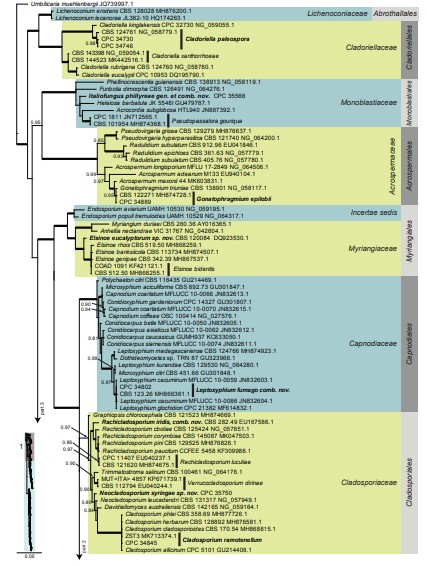
Fig. 3, parts 1–4. Consensus phylogram (50 % majority rule) resulting from a Bayesian analysis of the Dothideomycetes LSU sequence alignment. Bayesian posterior probabilities (PP) > 0.79 are shown at the nodes and the scale bar represents the expected changes per site. Thickened branches represent PP = 1. Families and orders are indicated with coloured blocks to the right of the tree. GenBank accession and/or culture collection numbers are indicated for all species. The tree was rooted to Umbilicaria muehlenbergii (strain A18, GenBank JQ739997.1) and the species treated in this study for which LSU sequence data were available are indicated in bold face. A miniature overview tree is also presented to facilitate navigation along the tree topology.
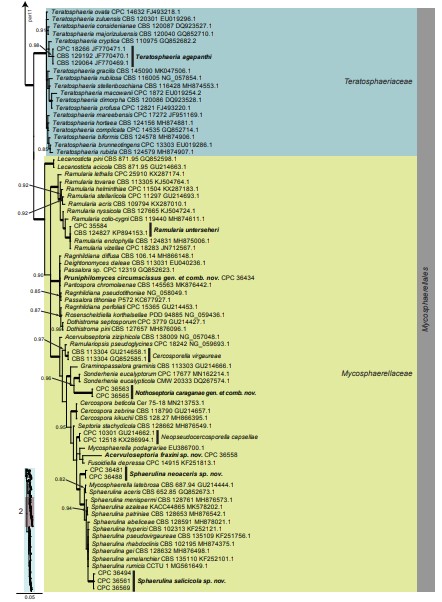
Fig. 3. (Continued).
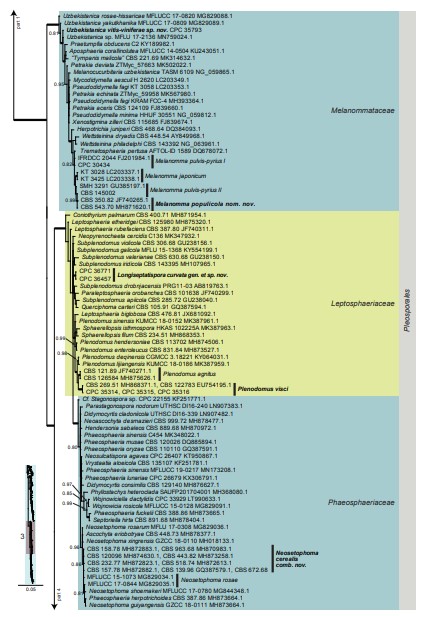
Fig. 3. (Continued).
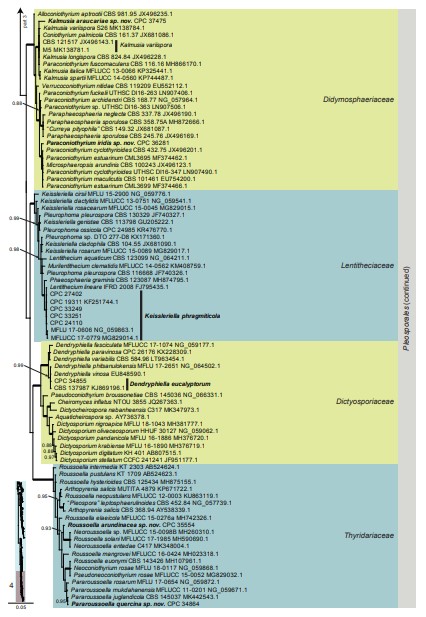
Fig. 3. (Continued).
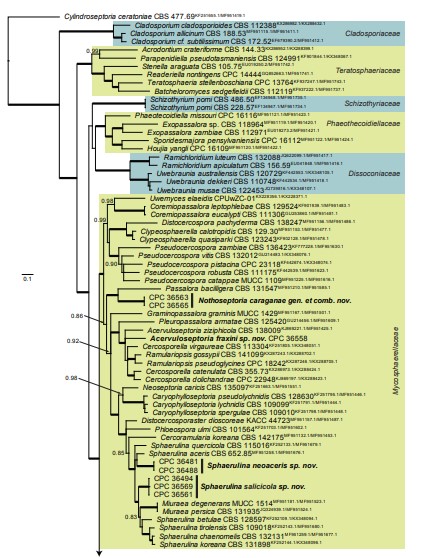
Fig. 8, parts 1, 2. Consensus phylogram (50 % majority rule) resulting from a Bayesian analysis of the Mycosphaerellaceae LSU/rpb2 sequence alignment (123 strains including the outgroup; 246 and 500 unique site patterns for LSU and rpb2, respectively; 35 928 sampled trees from 4 330 000 generations). The alignment is derived from the overview LSU/rpb2 alignment of Videira et al. (2017). Bayesian posterior probabilities (PP) > 0.79 are shown at the nodes and the scale bar represents the expected changes per site. Thickened branches represent PP = 1. Families are indicated with coloured blocks to the right of the tree. GenBank accession (superscript text) and/or culture collection numbers are indicated for all species. The tree was rooted to Cylindroseptoria ceratoniae (culture CBS 477.69; GenBank KF251655.1 and MF951419.1 for LSU and rpb2, respectively) and the species treated in this study are indicated in bold face.
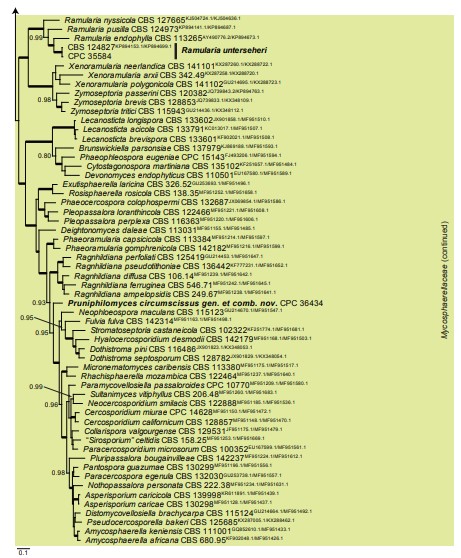
Fig. 8. (Continued)

Fig. 43. Nothoseptoria caraganae (CPC 36563). A, B. Leaf spots on Caragana arborescens. C. Immersed conidiomata with conidial cirrus. D. Conidiomata with conidial cirri on SNA. E–G. Conidiogenous cells giving rise to conidia. H. Conidia. Scale bars: D = 250 µm, all others = 10 µm.
