Neosorocybe pini Crous & Akulov, sp. nov.
MycoBank number: MB 835093; Index Fungorum number: IF 835093; Facesoffungi number: FoF; Fig. 42.
Etymology: Name refers to the host genus Pinus from which the species was isolated.
Conidiomata synnematous, consisting of conidiophores that are brown, smooth, multiseptate, branched, subcylindrical, up to 300 µm tall. Conidiogenous cells integrated on conidiophores, terminal and intercalary, giving rise to branched conidial chains via sympodial proliferation, 7–12 × 2.5–3.5 µm, hila truncate, 2 µm diam, not thickened nor darkened. Conidia subcylindrical, medium brown, smooth, guttulate, 0–1(–2)-septate, forming branched chains of disarticulating conidia; ramoconidia 10–20 × 2–3 µm; conidia 5–20 × 2–3 µm; hila 1.5–2 µm diam, not thickened nor darkened.
Culture characteristics: Colonies flat, spreading, slimy, with sparse aerial mycelium and smooth, lobate margin, reaching 7 mm diam after 2 wk at 25 ºC. On MEA, PDA and OA surface and reverse iron-grey.
Typus: Ukraine, Sumy region, Okhtyrka district, Klymentove village, NNP Hetmanskyi, on Pinus sylvestris (Pinaceae), fallen trunk, 3 Aug. 2018, A. Akulov, HPC 2548 = CWU (Myc) AS 6816 (holotype CBS H-24172, culture ex-type CPC 36628 = CBS 146085).
Notes: Neosorocybe pini is phylogenetically allied to Sorocybe, but appears to represent a distinct genus, for which the name Neosorocybe is introduced. Neosorocybe has synnemata with chains of pigmented, cylindrical conidia, with typical culture characteristics of Chaetothyriales, with slimy, iron-grey colonies on MEA and PDA.
Based on a megablast search of NCBI’s GenBank nucleotide database, only distant hits were obtained using the ITS sequence, e.g. to Capronia villosa (strain ATCC 56206, GenBank AF050261.1; Identities = 397/454 (87 %), 23 gaps (5 %)), Sorocybe oblongispora (as Sorocybe sp. JBT-2019, strain NB-795-1, GenBank MN114115.1; Identities = 419/487 (86 %), 26 gaps (5 %)), and Sorocybe resinae (strain DAOM 239134, GenBank EU030275.1; Identities = 412/480 (86 %), 21 gaps (4 %)). Closest hits using the LSU sequence are Sorocybe resinae (strain DAOM 239134, GenBank EU030277.1; Identities = 803/826 (97 %), 1 gap (0 %)), Sorocybe oblongispora (as Sorocybe sp. JBT-2019a, strain NB-795-1, GenBank MN114117.1; Identities = 828/852 (97 %), 1 gap (0 %)), and Lasallia pustulata (voucher 5 June 01 Wedin (UPS), GenBank AY300839.1; Identities = 821/852 (96 %), 3 gaps (0 %)) – also see Fig.4.
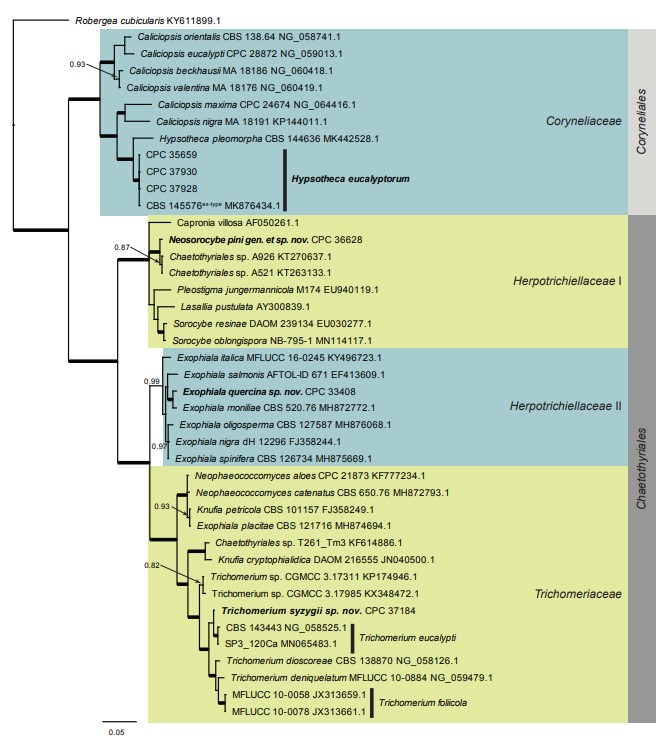
Fig. 4. Consensus phylogram (50 % majority rule) resulting from a Bayesian analysis of the Eurotiomycetes LSU sequence alignment. Bayesian posterior probabilities (PP) > 0.79 are shown at the nodes and the scale bar represents the expected changes per site. Thickened branches represent PP = 1. Families and orders are indicated with coloured blocks to the right of the tree. GenBank accession and/or culture collection numbers are indicated for all species. The tree was rooted to Robergea cubicularis (voucher G.M. 2013-05-09.1, GenBank KY611899.1) and the species treated in this study for which LSU sequence data were available are indicated in bold face.
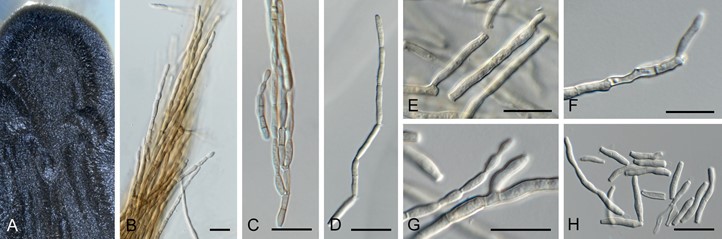
Fig. 42. Neosorocybe pini (CPC 36628). A. Colony on SNA. B. Synnema. C, D. Conidial chains. E–G. Conidiogenous cells. H. Conidia. Scale bars = 10 µm.
