Neoheleiosa lincangensis Mortimer sp. nov. Figure 4.
MycoBank number: MB 838518; Index Fungorum number: IF 838518; Facesoffungi number: FoF 11960;
Etymology: The specific epithet reflects Lincang, from where the holotype was collected.
Holotype: HKAS 111914
Habitat terrestrial, saprobic on dead twigs of Pittosporum sp. Sexual morph: Ascomata, 130–160 µm high, 200–250 µm diam. (x = 146 × 217 µm, n = 5), solitary, scattered, immersed to erumpent, globose to subglobose or obpyriform, dark brown to black, coriaceous, ostiolate. Ostiole 70–110 µm long, 30–50 µm diam. (x = 90 × 40 µm, n = 5), eccentric, papillate, black, smooth, filled with hyaline cells when mature. Peridium 8–12 µm thick at the base, 15–25 µm wide at sides, comprising blackish to dark brown, thin-walled cells of textura globulosa. Hamathecium 2–2.5 µm wide, comprising numerous, filamentous, branched, septate, pseudoparaphyses. Asci 115–135 × 10–12 µm (x = 124 × 10.6 µm, n = 30), 8-spored, bitunicate, fissitunicate, cylindrical, pedicel furcate, rounded and thick-walled at the apex, with an ocular chamber. Ascospores 16.5–17.5 × 7–9 µm (x = 17.1 × 8.2 µm, n = 30), overlapping uniseriate, narrowly ovoid to clavate, 1-septate, initially hyaline, becoming dark brown at maturity, with conically rounded ends, guttulate, thick walled, faintly longitudinally striated, lacking a mucilaginous sheath. Asexual morph: Undetermined.
Material Examined: China, Yunnan Province, Lincang, Yongde County, Bankaxiang, (N: 23.997479, E: 99.480670), on dead twigs of Pittosporum sp., 8 April 2019, D.N. Wanasinghe, DW0738-07 (HKAS 111914, holotype); ibid. DW0738-09 (HKAS 111911); DW0738-11 (HKAS 111912); DW0738-12 (HKAS 111913).
Notes: Four specimens with 1-septate, brown ascospores species were collected on dead twigs of Pittosporum from Bankaxiang in Yunnan Province. We made several attempts to obtain a culture via single spore isolations and direct isolation from fungal tissues (Senanayake et al., 2020). However, we were unable to get a pure culture, and DNA was extracted directly from the fruiting bodies. Sequence data of these four collections grouped in a strongly supported monophyletic clade in the concatenated SSU, LSU, ITS, tef 1 and mtSSU sequence analyses (Figure 1). Morphologically all these collections were identical and there were no differences in their DNA sequence data. Thus, we recognize that all of these specimens belong to a single species, Neoheleiosa lincangensis sp. nov.
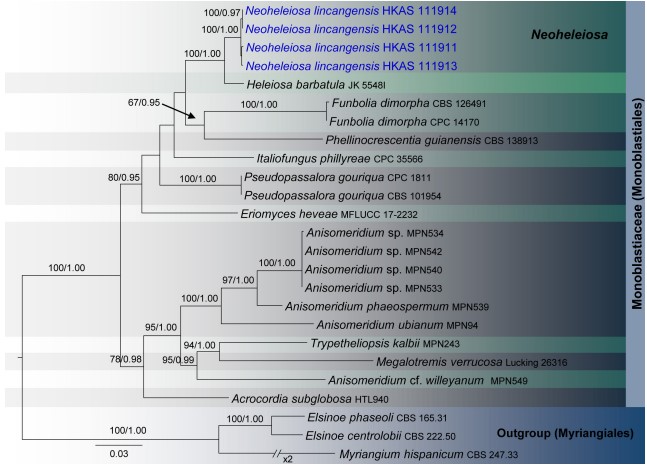
FIGURE 1 | RAxML tree based on a combined dataset of partial SSU, LSU, ITS, tef 1, and mtSSU sequence analyses in Monoblastiaceae. Bootstrap support values for ML equal to or greater than 60%, Bayesian posterior probabilities (BYPP) equal to or greater than 0.95 are shown as ML/BI above the nodes. The new isolates are in blue. The scale bar represents the expected number of nucleotide substitutions per site.

FIGURE 4 | Neoheleiosa lincangensis (HKAS 111914). (a) Ascomata observed on host substrate. (b,d) Horizontal sections of ascomata. (c) Vertical sections through an ascoma. (e,f) Cells of peridium. (g) Pseudoparaphyses. (h–j) Asci. (k) Ocular chamber. (l) Pedicel. (m–q) Ascospores (note: q longitudinally striated). Scale bars: (c,d) = 100 µm, (e, f, k–q) = 10 µm, (g) = 5 µm, (h–j) = 20 µm.
