Neoxylomyces multiseptatus MS Calabon, Boonmee, E.B.G. Jones & K.D. Hyde, sp. nov. (Figure 8)
MycoBank number:MB 838555; Index Fungorum number: IF 838555; Facesoffungi number: FoF 09534;
Etymology: In reference to the multiseptate chlamydospores
Holotype: MFLU 21–0014
Saprobic on decaying wood submerged in freshwater habitats. Sexual morph: Undetermined. Asexual morph: Colonies on the substrate effuse, scattered, dark brown to black. Mycelium mostly immersed, composed of branched, septate, smooth, thin-walled, dematiaceous, anastomosing hyphae. Conidiophores and conidia not developed. Chlamy- dospores 111–378 × 8–13 µm (x = 255 × 10.2 µm, n = 30), narrowly fusiform, cylindrical, intercalary, erect, mostly straight, slightly curved, solitary or in chains, occasionally branched, with 7–45 septa, constricted at septa, brown, paler and truncated end cells, 3.28–5.93 µm wide (x = 4.39 µm), thick-walled with scarce irregular longitudinal striations.
Culture characteristics: Chlamydospores germinating on malt extract agar (MEA) within 24 h. Germ tubes produced from the basal and apical cell of conidia. Colonies growing on MEA, reaching 35–40 mm in 4 weeks at 25 ◦C. Mycelia superficial, circular, with entire margin, flat, smooth, from above ivory to pale brown at the margin, white at the center; reverse, dark brown at the center then becoming pale orange to light brown at the margin.
Material examined: THAILAND, Chiang Mai Province, Mae Teang District, Mushroom Research Center (M.R.C.), on decaying wood submerged in a stream, 11 February 2019, M.S. Calabon, MC02 (MFLU 21–0014, holotype), ex-type living culture, MFLUCC 21–0007.
GenBank accession numbers: LSU = MW287236, SSU = MW287239, ITS = MW260332,TEF1-α = MW512607
Notes: Neoxylomyces multiseptatus is similar to Xylomyces giganteus in having brown, long, multiseptate chlamydospores. The former differs in the number of septa (7–45 versus 6–85), size of the chlamydospores (111–378 × 8–13 µm versus (140) 190–575 × 25–50 µm), and absence of a mucilaginous coating to the chlamydospores [69,70]. Furthermore, Xylomyces belongs to Aliquandostipitaceae (Jahnulales, Dothideomycetes) [49]. The closest match of the sequences based on BLASTn searches in NCBI GenBank database were Clathrosporium. Clathrosporium retortum (CCIBt 4123) was the closest species based on BLAST result of ITS (83% similarity) and LSU sequence data (95% similarity). The TEF1- α sequence was 91% similar to Tolypocladium ophioglossoides (NBRC:8992), T. paradoxum (NBRC:106958), Metarhizium granulomatis (UAMH 11176), and Hypomyces polyporinus (ATCC 76479). The multi-loci phylogenetic analyses show that N. multiseptatus is a distinct species and sister taxon to Clathrosporium retortum (CCIBt 4123) with 100% MP, 1.00 BYPP support. The former has long, narrowly fusiform, multiseptate chlamydospores while the latter has subglobose to irregular, hyaline to subhyaline conidia formed by branched, densely interwoven conidial filaments [71].
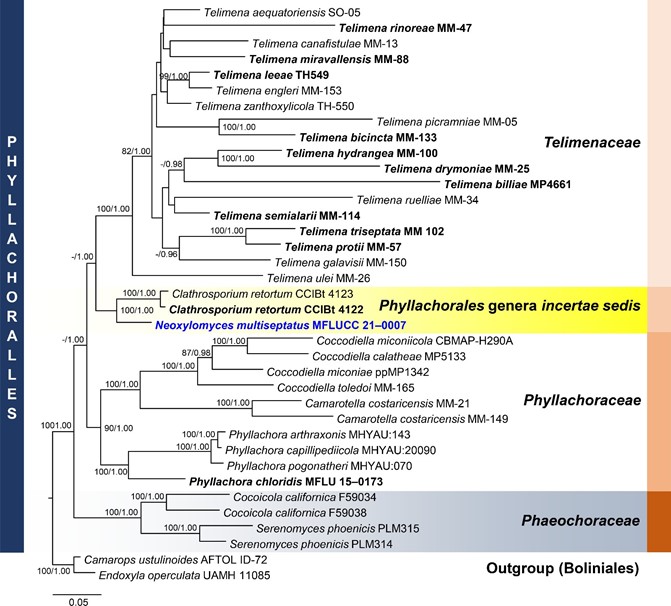
Figure 8. Phylogram generated from maximum likelihood analysis based on combined LSU, SSU, ITS, and TEF1-α sequence data representing the species of Phyllachorales and related taxa. Sequences are taken from Dayarathne et al. [72] and Yang et al. [73]. Thirty-seven taxa were included in the combined analyses, which comprised 3259 characters (LSU = 863, SSU = 1175, ITS = 462, TEF1-α = 759) after alignment. The best scoring RAxML tree with a final likelihood value of −32,589.920351 is presented. The matrix had 2136 distinct alignment patterns, with 34.35% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.248688, C = 0.249512, G = 0.274933, T = 0.226867; substitution rates: AC = 1.044099, AG = 1.599779, AT = 1.069911, CG = 1.048973, CT = 3.108380, GT = 1.000000; gamma distribution shape parameter α = 0.745075. Bootstrap support values for ML equal to or greater than 75% and BYPP equal to or greater than 0.95 are given above the nodes. Camarops ustulinoides (AFTOL-ID 72) and Endoxyla operculata (UAMH 11085) were used as the outgroup taxa. The newly generated sequence is indicated in blue. The ex-type strains are indicated in bold.
