Massaria racemosa Samarak. & K.D. Hyde, sp. nov. Figure 3
MycoBank number: MB 843666; Index Fungorum number: IF 843666; Facesoffungi number: FoF 10808;
Etymology: The specific epithet refers to the host species racemosa.
Holotype: MFLU 19–2135
Saprobic on a dead branch. Sexual morph: Ascomata 380–450 μm diam. × 290–320 μm high (M = 420 × 307 μm, n = 10), immersed in bark with erumpent neck, visible as black dots on the host surface, solitary, scattered or sometimes gregarious, sub globose, flat base, coriaceous, brown to dark brown, with centrally opening ostiole. Ostiole centrally located, filled with periphyses and hyaline cells. Peridium 18–28 μm (M = 24 μm, n =12) wide at the base, 42–70 μm (M = 55.5 μm, n =8) wide around the ostiole, outer layer comprising reddish to dark brown, fused with host tissues, thin-walled cells of textura angularis, inner layer composed of hyaline, loosen, cells of textura angularis. Hamathecium 1.4–3 μm (M = 2.1 μm, n = 15) wide, composed of numerous, dense, long, filamentous, branched, septate, trabeculate, round to blunt apex, pseudoparaphyses, embedded in a gelatinous matrix. Asci 175–260 × 22–42 μm (M = 215 × 32 μm, n = 25), 8-spored, bitunicate, fissitunicate, cylindrical or oblong, with a short pedicel, apically flattened. Ascospores 45–56 × 11–16 μm (M = 50 × 14 μm, n = 35), overlapping 2–3 seriate, fusoid to ellipsoid, hyaline, many guttules when immature, 3 septate when mature, deteriorated dark brown when over mature, lumped at the septum, narrowly rounded to nearly acute at the ends, rough-walled, prominent, large rhomboid guttule in each cell, surrounded by a 9–28 μm (M = 15 μm, n = 10) wide, distinct mucilaginous sheath. Asexual morph: Undetermined.
Material examined: Thailand, Pua district, Nan province, on a dead branch of Ficus racemosa (Moraceae) attached to the host, 29 January 2019, MC. Samarakoon, SAMC215 (MFLU 19-2135, holotype), isotype HKAS 106992. Additional sequences ITS XXXX, TUB2 XXXX.
Notes: Massaria racemosa morphology fits into the generic concept of Massaria as above-mentioned and differs from M. broussonetiae as having sub globose, flat base ascomata, lacking green algae-like structures, and hyaline ascospores. The LSU sequence of M. racemosa is similar to that of M. parva WU 30553 (844/850; 99%), M. lantanae CBS 125592 (840/851; 99%), and M. anomia WU 30503 (840/851; 99%), while SSU is similar to that M. lantanae CBS 125592 (786/794; 99%), M. anomia WU 30509 (786/794; 99%), and M. ulmi WU 30566 (782/794; 98%). The RPB2 sequence of M. racemosa is similar to that of M. anomia CBS 591.78 (901/1032; 87%), M. inquinans WU 30527 (899/1039; 87%), M. vomitoria WU 30606 (897/1038; 86%), and M. platanoidea WU 30554 (898/1039; 86%). Combined gene phylogenies also show that M. racemosa clusters as a distinct clade, and here we introduce it as a new species.
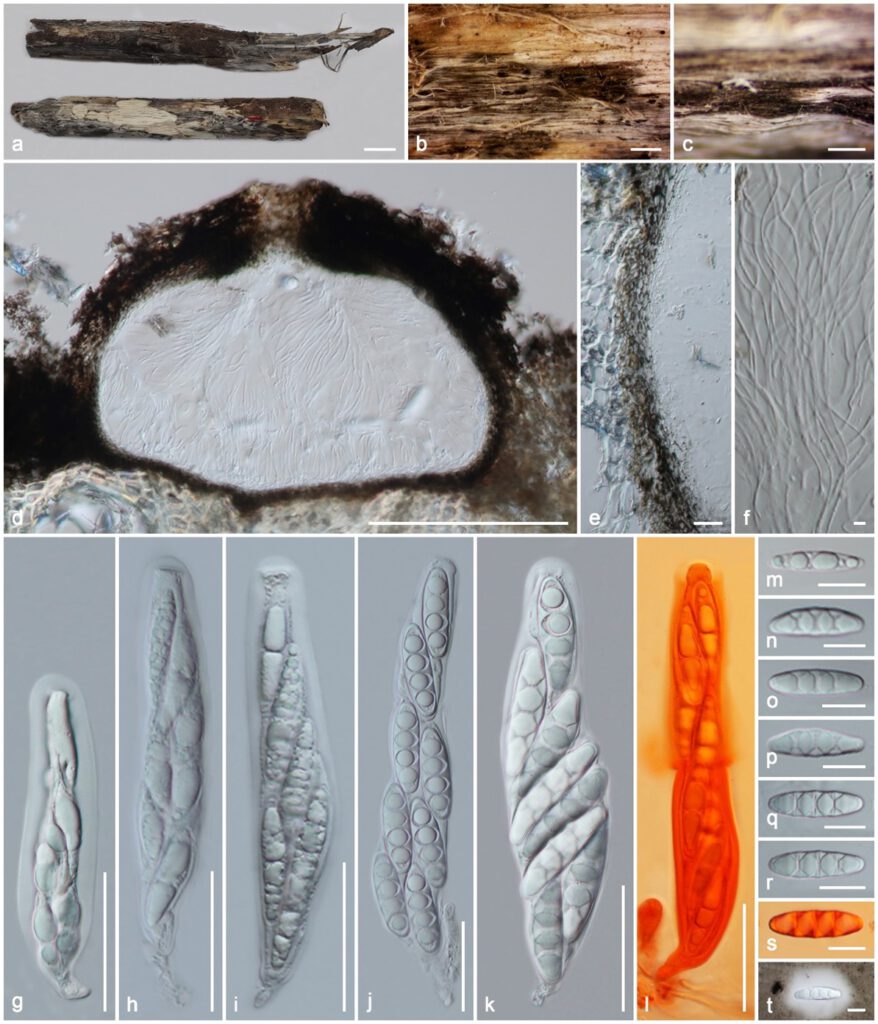
Figure 3. Massaria racemosa (holotype MFLU 19–2135). (a) Substrate; (b,c) ascomata on substrate; (d) vertical section of ascoma; (e) peridium; (f) paraphyses; (g–l) asci (l in Congo Red); m–t ascospores (s in Congo Red, t in Indian ink). Scale bars: (a) = 1 cm; (b) = 1000 µm; (c) = 500 µm; (d) = 200 µm; (g–l) = 50 µm; (e,m–s) = 20 µm; (f) = 5 µm.
