Iodosphaeriaceae O. Hilber, The Genus Lasiosphaeria and Allied Taxa (Kelheim): 7 (2002)
MycoBank number: MB 82138; Index Fungorum number: IF 82138; Facesoffungi number: FoF 06189; 8 species.
Saprobic on dead twigs and leaves of various hosts. Sexual morph: Ascomata superficial, solitary, black, and easily removed from the substrate, covered with dark brown, setae-like, brown hairs, comprising agglutinated mycelial strands, with a stellate arrangement, arising from cells at the perithecial surface. Ostioles pore-like opening, central, with periphyses. Peridium outer region comprising pigmented, brown cells of textura angularis, inner region comprising hyaline, flattened cells. Paraphyses numerous, hypha-like, septate, flexuose, slightly tapered towards the apex. Asci 8-spored, unitunicate, narrowly clavate, short pedicellate or apedicellate, apex rounded, with a J+, subapical ring. Ascospores biseriate, hyaline, allantoid, unicellular, smooth-walled, lacking sheaths or appendages. Asexual morph: ceratosporium-like conidia have been observed on the surface of perithecia, but may not be related.
Type genus – Iodosphaeria Samuels, E. Müll. & Petrini
Notes – Iodosphaeriaceae was introduced to accommodate Iodosphaeria and placed in Amphisphaeriales (Hilber & Hilber 2002). Maharachchikumbura et al. (2016b) and Samarakoon et al. (2016) placed this family within Xylariales. Iodosphaeriaceae was placed in Xylariomycetidae, families incertae sedis (Hyde et al. 2017b, Hongsanan et al. 2017, Wijayawardene et al. 2018a). We accept Iodosphaeriaceae in Amphisphaeriales based on multigene analysis of combined LSU, ITS, rpb2 and tub2 sequence data (Fig. 5).
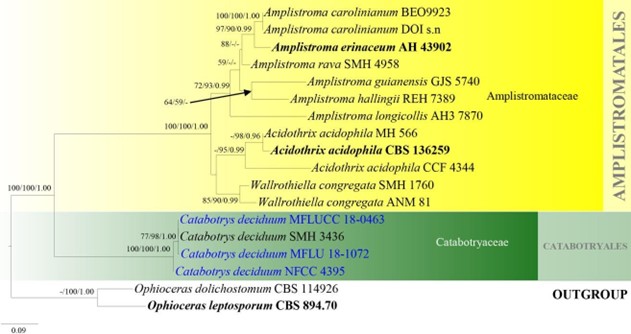
Figure 5 – Phylogram generated from maximum likelihood analysis based on combined LSU and ITS sequence data of Amplistromatales. Eighteen strains are included in the combined analyses which comprised 1661 characters (908 characters for LSU, 753 characters for ITS) after alignment. Ophioceras dolichostomum (CBS 114926) and O. leptosporum (CBS 894.70) are used as outgroup taxa. Single gene analyses were also carried out and the phylogenies were similar in topology and clade stability. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RaxML tree with a final likelihood value of – 6986.920872 is presented. Estimated base frequencies were as follows: A = 0.226, C = 0.264, G = 0.321, T = 0.190; substitution rates AC = 0.643764, AG = 1.471407, AT = 1.612417, CG = 0.654293, CT = 5.330856, GT = 1.000000; gamma distribution shape parameter a = 0.653797. Bootstrap support values for MP and ML greater than 55% and Bayesian posterior probabilities greater than 0.95 are given near the nodes. Ex-type strains are in bold. The newly generated sequences are indicated in blue.
Genera
