Gonatobotrys Corda, Pracht-Fl. Eur. Schimmelbild.: 9 (1839)
MycoBank number: MB 8374; Index Fungorum number: IF 8374; Facesoffungi number: FoF14485;
Saprobic on wood or vegetation. Sexual morph: Undetermined. Asexual morph: Hyphomycetous. Mycelium composed of hyaline to light brown, superficial, effused, smooth-walled, branched, septate hyphae. Conidiophores mononematous, macronematous, hyaline, erect, septate, branched or unbranched, smooth-walled. Conidiogenous cells terminal to intercalary, polyblastic, globose to subglobose, hyaline to pale brown, swollen. Conidia solitary, obovoid to ellipsoidal, with a raised rim at base, multi-septate, holoblastic, hyaline to pale brown, smooth to verrucose, conidial secession with conspicuous denticles on the conidia and conidiogenous cells (adapted from Hoch 1977, Walker & Minter 1981, Whaley & Barnett 1963).
Notes – Gonatobotrys was introduced by Corda (1839), and is well known as fungicolous hyphomycetes of Alternaria and Fusarium species (Hoch 1977, Walker & Minter 1981, Whaley & Barnett 1963). The type species Gonatobotrys simplex was considered as the asexual morph of Melanospora damnosa, and was synonymised as M. simplex (Vakili 1989, Réblová et al. 2016). However, Crous et al. (2020b) proposed that characters of hyphomycetous Gonatobotrys were never observed in Melanospora cultures. Melanospora damnosa is distinct from G. simplex based on phylogenetic results (Vu et al. 2019, Crous et al. 2020b). In this study, Gonatobotrys is sister to Vittatispora (56%ML/0.97BY) in Ceratostomataceae, but distinct from Melanospora clade based on multi-gene phylogeny (Fig. 1). Therefore, we accept Gonatobotrys as a genus in Ceratostomataceae.
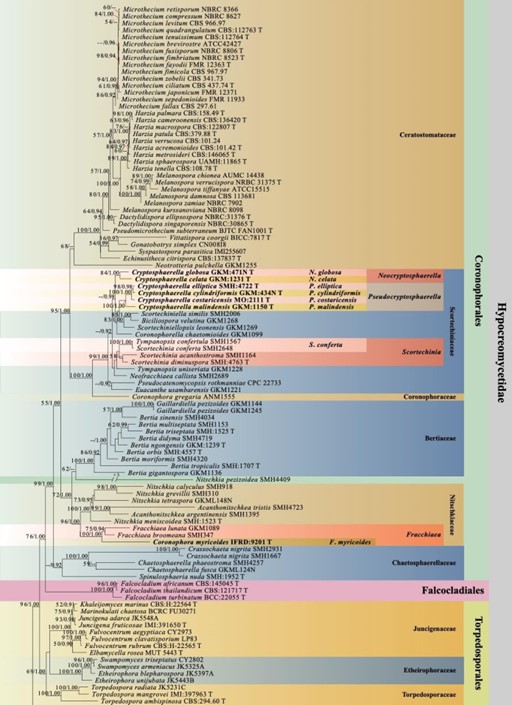
Figure 1 – Phylogram generated from maximum likelihood analysis based on combined LSU, SSU, TEF, RPB2, TUB and ITS sequence data with the confidence values of bootstrap (BS) proportions from the Maximum Likelihood (ML) analysis (ML-BS > 50%, before the backslash) and the posterior probabilities (PP) from the Bayesian (BY) analysis (BY-PP > 0.90, after the backslash) above corresponding nodes. The ‘–’ indicates a lack of statistical support (< 50% for ML-BS and < 0.90 for BY-PP). One hundred and ninety-eight strains are included in the combined analyses which comprised 4581 characters (842 characters for LSU, 1025 characters for SSU, 950 characters for TEF, 876 characters for RPB2, 391 characters for TUB, 497 characters for ITS) after alignment. Strains of Xylariomycetidae are used as outgroup taxa. The best score in IQ-TREE explores with a final likelihood value of -90984.2293 is presented. The model of each partitioned gene is LSU: GTR+I+G; SSU: GTR+G; TEF: GTR+I+G; RPB2: GTR+I+G; TUB: GTR+I+G; ITS: GTR+G. Sequences generated indicated in bold. Strain numbers are noted after the species names and ex-type strains marked with ‘T’ after the culture number. Alignment is available at TreeBASE (URL: http://purl.org/phylo/treebase/phylows/study/TB2:S28271).
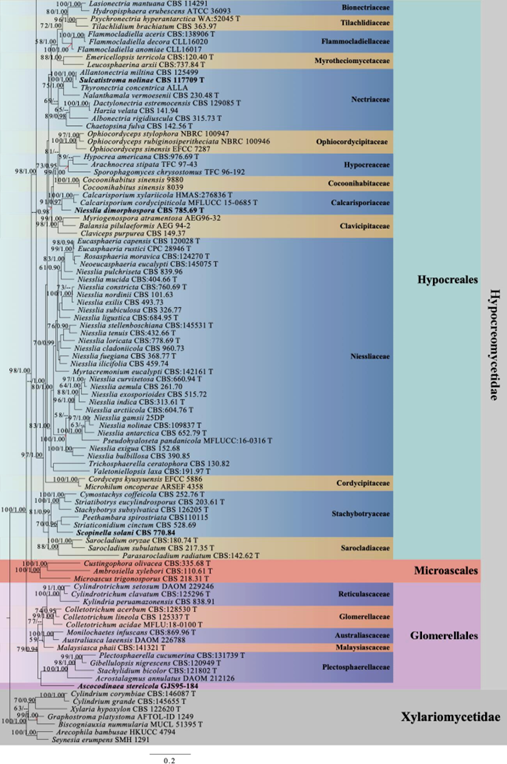
Figure 1 – Continued.
Species
