Eriocamporesia R.H. Perera, Samarak. & K.D. Hyde, gen. nov.
Index Fungorum number: IF556791; Facesoffungi number: FoF 06959
Etymology: Named after the collector Erio Camporesi
Associated with twigs of dicotyledonous plant. Sexual morph Ascostromata on host single, appearing as yellow pustules, pulvinate, semi-immersed in plant tissue, orange, KOH + purple; ascostromatic tissue pseudoparenchymatous, covering the top of the ascomatal bases. Ascomata perithe- cial, usually valsoid, embedded in stromata at irregular lev- els, fuscous black, necks emerge at stromatal surface as dark orange brown ostioles covered with orange stromatal tissue to form papillae extending above stromatal surface. Peridium of 5–7 layers of flattened, elongate, brown-walled cells of textura angularis. Paraphyses lacking. Asci 8-spored, uni- tunicate, oblong ellipsoidal to fusoid to ellipsoidal, apedi- cellate, floating freely in the perithecial cavity, thin-walled, apex simple. Ascospores 2-seriate, cylindrical to allantoid, occasionally ellipsoidal, ends round, curved or sometimes straight, aseptate, hyaline, smooth-walled, without append- ages. Asexual morph Conidiostromata appearing as yellow pustules on the host, occur as separate structures, pulvinate, semi-immersed, orange. Conidiomata multi-locular, with locules often convoluted, apapillate, with 1-ostiolar opening, orange, KOH + purple; stromatic tissue pseudoparenchyma- tous. Paraphyses lacking. Pycnidial walls of 2–4 layers of flattened, elongate, hyaline, cells of textura angularis. Con- idiophores cylindrical, rarely septate or branching, hyaline. Conidiogenous cells phialidic, cylindrical with attenuated apices, hyaline. Conidia hyaline, cylindrical or allantoid, ends round, aseptate, hyaline, smooth-walled, without appendages.
Type species: Eriocamporesia aurantia R.H. Perera, Samarak. & K.D. Hyde
Notes: DNA sequence evidence shows Eriocamporesia is a member of Cryphonectriaceae and distinct from other genera in the family (Fig. 108). Eriocamporesia resembles other genera in Cryphonectriaceae by orange ascostromata and conidiostromata which turned purple in 3% KOH, and phialidic conidiogenous cells (Gryzenhout et al. 2006; Jiang et al. 2019b). Eriocamporesia shares similar morphology with Holocryphia in having orange stromata, valsoid, fuscous black ascomata and apedicellate asci (Chen et al. 2013a). However, it can be segregated from Holocryphia by the lack of paraphyses in the conidiomata and allantoid conidia, while Holocryphia has paraphyses and cylindrical conidia. Phylogenetic analyses based on a combined LSU, ITS and TUB2 sequence dataset shows that Eriocamporesia forms a distinct lineage basal to the genera Endothia and Aurantioporthe G.L. Beier & Blanchette in Cryphonectri- aceae. Considering both molecular and morphological data we introduce Eriocamporesia as a new genus.
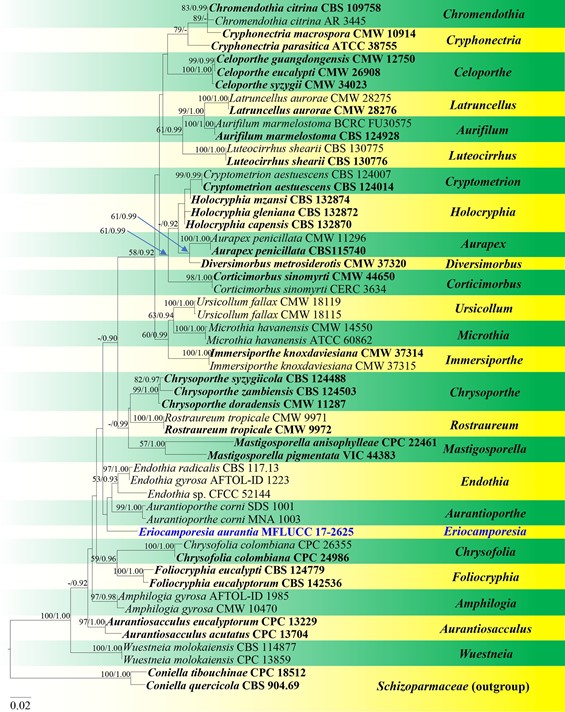
Fig. 108 Phylogram generated from maximum likelihood analysis based on combined LSU, ITS and TUB2 sequence data representing Cryphonectriaceae. Related sequences are taken from Senanayake et al. (2017). Fifty-four strains are included in the combined analyses which comprise 1830 characters (866 characters for LSU, 639 char- acters for ITS, 323 characters for TUB2) after alignment. Coniella tibouchinae (CPC 18512) and C. quercicola (CBS 904.69) in Schizo- parmaceae is used as the outgroup taxa. Single gene analyses were also performed to compare the topology and clade stability with com- bined gene analyses. Tree topology of the maximum likelihood analy- sis is similar to the Bayesian analysis. The best RAxML tree with a final likelihood values of − 7862.571574 is presented. The matrix had 525 distinct alignment patterns, with 29.64% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.239368, C = 0.254757, G = 0.280128, T = 0.225747; substitution rates AC = 2.087607, AG = 3.025115, AT = 2.060161, CG = 1.371584, CT = 7.165898, GT = 1.000000; gamma distribution shape parameter α = 0.727546. Bootstrap values for maximum likelihood (ML) equal to or greater than 50% and clade credibility values greater than 0.90 (the rounding of values to 2 decimal proportions) from Bayesian- inference analysis labelled on the nodes. Ex-type strains are in bold and black, the new isolate is indicated in bold and blue
Species
