Discosia celtidis Tennakoon, C.H. Kuo & K.D. Hyde, Fungal Diversity 108, 108 (2021)
Index Fungorum number: IF 555455; Facesoffungi number: FoF 09345; Fig. 1
Saprobic on the dead leaves of Trema orientale. Sexual morph: Undetermined. Asexual morph: Coelomycetous. Conidiomata 50–100 × 250–400 µm (x̅ = 71 × 348 µm, n = 10), conspicuous, pycnidial, stromatic, amphigenous, scattered or aggregated, black, flattened or concave at the centre with a convex margin and a relatively thin stromatic base, rounded, glabrous, epidermal, unilocular or multilocular. Conidiomata wall 10–15 μm wide, thin-walled, composed of several layers of small, flattened, brown to dark brown pseudoparenchymatous cells, cells towards the inside light brown, arranged in a textura angularis, with outer layers intermixed with the host tissues. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 7–10 × 1–3 µm (x̅ = 8.7 × 2.4 μm, n = 20), holoblastic to phialidic, ampulliform, integrated, hyaline, smooth-walled. Conidia 18–25 × 3–4 μm (x̅ = 23 × 3.5 μm, n = 40), hyaline, fusiform to cylindrical, thick-walled, smooth, straight or slightly curved, 3-septate, slightly constricted at one septum at apex, with cells of equal width, basal cell obconic, with a truncate base, apical cell subconical with a rounded apex, unbranched, filiform, flexuous or straight appendage; presence of both ends, apical and basal appendages 10–12 μm (x̅ = 10.8 μm).
Culture characteristics – Colonies on PDA, 20–25 mm, diam. after 2 weeks, colony from above medium dense, raised, surface smooth with entire edge, fluffy to velvety with smooth aspects, light brown to dark brown at the margin, dark brown to dark grey in the centre; colony from below light brown at the margin, dark brown to black in the centre, pigmentation not produced in media.
Material examined – China, Taiwan Province, Chiayi, Fanlu Township area, Dahu Forest, dead leaves of Trema orientale, 25 August 2019, D.S. Tennakoon, TAP015 (MFLU 18-2604), living culture MFLUCC 19-0132.
GenBank accession numbers – LSU: OP647860, ITS: OP650239.
Known distribution (based on molecular data) – China (Tennakoon et al. 2021a, this study).
Known hosts (based on molecular data) – Celtis formosana (Tennakoon et al. 2021a), Trema orientale (this study).
Notes – In this study, our isolate (MFLUCC 19-0132) shares similar morphological characteristics with Discosia celtidis in having hyaline, fusiform to cylindrical conidia with apical and basal appendages (Tennakoon et al. 2021a). In particular, they have conidia (19–24 × 3–4 μm vs. 18–25 × 3–4 μm) and conidiogenous cells (7–9 × 1–3 µm vs. 7–10 × 1–3 µm) of overlapped size ranges. Phylogeny also shows that our collection is grouped with Discosia celtidis isolates with 96% maximum likelihood bootstrap support and 0.98 Bayesian posterior probability (Fig. 76). Therefore, we introduce our isolate as a new host record of Discosia celtidis from Trema orientale (Cannabaceae). Discosia celtidis was initially introduced by Tennakoon et al. (2021a) from Celtis formosana (Cannabaceae). Interestingly, this record was also collected from another species from the same family.
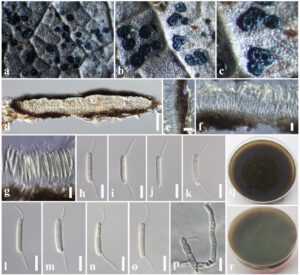
Fig. 1 – Discosia celtidis (MFLU 18-2604, a new host record). a Appearance of conidiomata on the host. b, c Close-up of conidiomata. d Section of a conidioma. e Conidioma wall. f, g Conidiogenous cells. h–o Conidia. p Germinated conidium. q Colony from above. r Colony from below. Scale bars: d = 50 µm, e–p = 10 µm.

Fig. 2 – Phylogram generated from maximum likelihood analysis based on combined LSU and ITS sequence data. Forty-nine strains are included in the combined analyses which comprised 1447 characters (879 characters for LSU and 568 characters for ITS) after alignment. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RaxML tree with a final likelihood value of – 4353.810124 is presented. The matrix had 315 distinct alignment patterns with 5.74 % of undetermined characters or gaps. The evolutionary model GTR+I+G is applied to LSU and ITS genes. Bootstrap support values for ML equal to or greater than 70% and Bayesian posterior probabilities equal to or greater than 0.95 are given near nodes, respectively. The tree is rooted with Pestalotiopsis versicolor (BRIP 14534). Ex-type strains are in bold. The newly generated sequences are indicated in yellow.
