Colletotrichum godetiae Neerg., Friesia 4(1-2): 72 (1950) [1949-50], (KIB4) (Ruvi)
MycoBank number: MB 295335; Index Fungorum number: IF 295335; Facesoffungi number: FoF 03881; Fig. **.
Saprobic on dead stems of Rhododendron sp. Sexual morph: Undetermined. Asexual morph: Vegetative hyphae 1–7 μm diam, hyaline to pale brown, smooth-walled, septate, branched. Chlamydospores not observed. Conidiomata absent, conidiophores formed directly on hyphae. Setae not observed. Conidiophores hyaline, smooth-walled, simple, to 14 μm long. Conidiogenous cells hyaline, smooth-walled, cylindrical, often with only short necks, 4–14 × (1.5–)3–6 μm, opening 1.5–2 μm diam, collarette 0.5 μm long, periclinal thickening observed. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical to fusiform with both ends acute or one end round and one end slightly acute, (7–)10.5–14.5(–15.5) × (3.5–)4–5(–5.5) μm, mean ± SD = 12.4 ± 2.0 × 4.3 ± 0.5 μm, L/W ratio = 2.9, strains CBS 127561, CBS 129917, CBS 193.32 and CBS 129951 differ in forming cylindrical to clavate conidia with one round and one acute end, conidia of strain CBS 862.70 are larger, measuring (8–)14–19(–24) × (4–)4.5–5(–5.5) μm, mean ± SD = 16.4 ± 2.4 × 4.9 ± 0.4 μm, L/W ratio = 3.4. Appressoria solitary, medium brown, smooth-walled, clavate to elliptical, the edge entire or undulate (8–)9–12.5(–14.5) × (3–)4–5.5(–6) μm, mean ± SD = 10.7 ± 1.9 × 4.7 ± 0.7 μm, L/W ratio = 2.3.
Culture characteristics – On PDA, Colonies on SNA flat with entire margin, hyaline, with little low white aerial mycelium, on Anthriscus stem growth rate 21–21.5 mm in 7 d (30.5–31.5 mm in 10 d). Colonies on OA flat with entire margin; surface salmon to hazel, no aerial mycelium, reverse salmon to vinaceous buff; growth rate 21–24 mm in 7 d (30–33.5 mm in 10 d). Conidia in mass not observed in strain CBS 133.44, but in strain CBS 125972 orange.
Material examined – China, Yunnan Province, Kunming distric, on dead leaves of Rhododendron sp., 23 August 2019, Napalai Chaiwan, KIB4 (MFLUCC****, living culture).
Host – Aeschynomene virginica, seed of Clarkia (syn. Godetia), rotten fruit of Olea europaea, Fragaria × ananassa, Podocarpus sp., Sambucus nigra, Solanum betaceum, Ugni molinae (Damm et al. 2012), Rhododendron sp.—(This study).
Distribution – Chile, Colombia, Denmark,Greece, Italy, Netherlands, Mexico, Schinus molle, South Africa (Damm et al. 2012), Thailand (Hyde et al. 2020), China — (This study).
GenBank accession numbers – MFLU **; ITS: ***; LSU: ***
Notes – Colletotrichum godetiae is a strain recorded from the different host of Colletotrichum godetiae (Damm et al. 2012). Colletotrichum godetiae also occurs on Rhododendron in Sweden and Latvia (Vinnere et al. 2002) same as with our isolation. The name of this species is contribute from the shape of the conidia (Faedda et al. 2011). The isolates from Rhododendron in Sweden and Latvia also showed the mainly clavate conidia (Vinnere et al. 2002). This species belongs to the C. gloeosporioides species complex. However, In a BLASTn search of GenBank, the ITS sequence had 99.82% similarity with Colletotrichum godetiae, the GAPDH sequence had 100% similarity with Colletotrichum godetiae, the TUB sequence had 99.64% similarity with Colletotrichum godetiae, while the ACT sequence had 100% similarity with Colletotrichum godetiae.
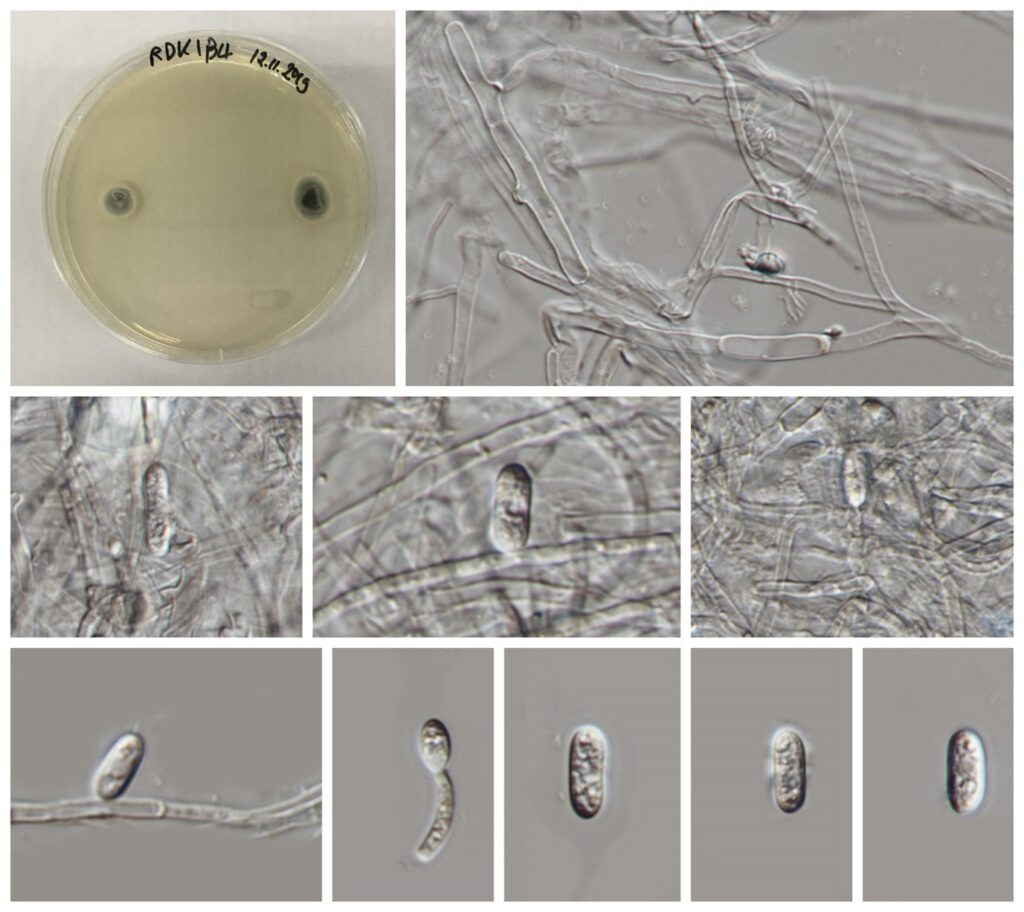
Figure***– Colletotrichum godetiae (MFLU **** KIB4). a, b Fruiting body on dead leaf of Dracaena sp. c–i Conidiophore. j–k Conidiogenous cells l–p Conidia. Scale bars: a = 500 , b = 200 μm, c–i = 20 μm, j–k = 10 μm, l–p = 5 μm.
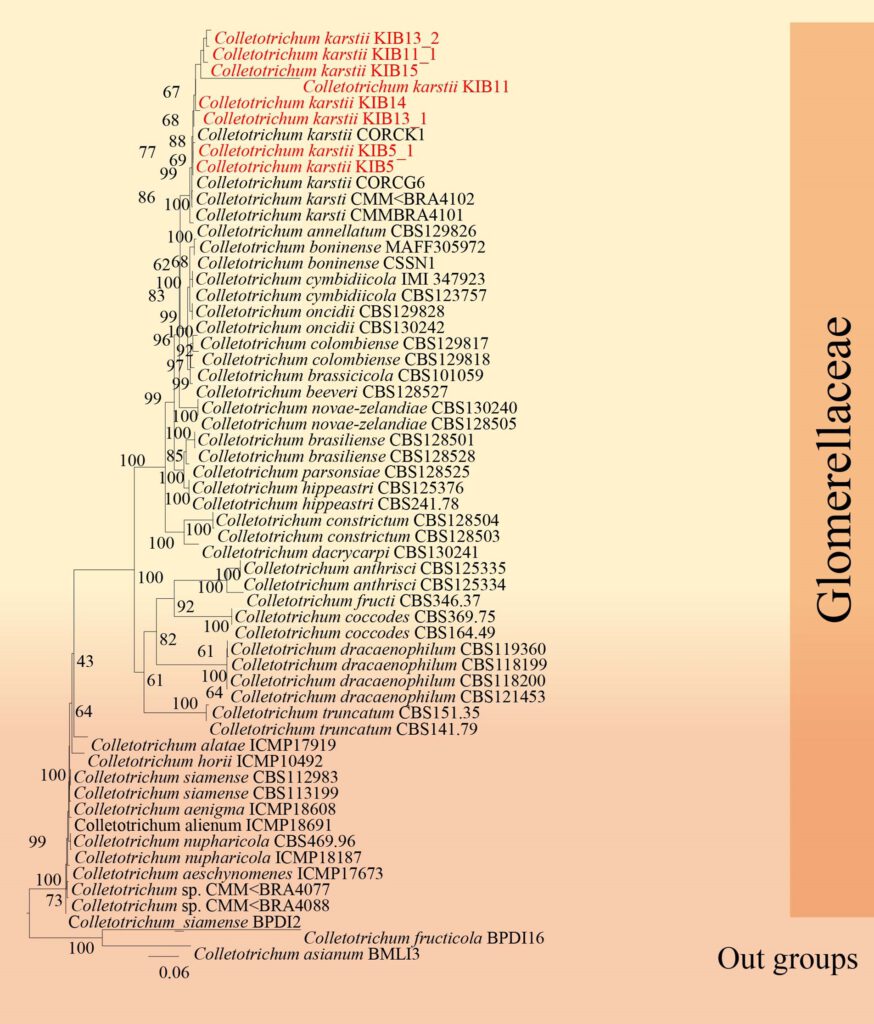
Figure**– Phylogram generated from RAxML analysis based on ITS, GAPDH, CHS, ACT and TUB sequence data of selected Glomerellaceae isolates. Related sequences were obtained from GenBank. Twenty-four taxa are included in the analyses, which comprise 1371 characters including gaps. The tree is rooted to Colletotrichum asianum. The best scoring RAxML tree with a final likelihood value of − 9826.525509 is presented. The matrix had 851 distinct alignment patterns, with 25.59% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.225440, C = 0.303246, G = 0.239417, T = 0.231897; substitution rates AC = 0.898292, AG = 2.455306, AT = 1.113846, CG = 0.871897, CT = 2.811180, GT = 1.000000; gamma distribution shape parameter α = 1.243027. Maximum likelihood bootstrap support values ≥ 75% (BT) are given at the nodes as ML. The scale bar indicates 0.04 changes. The isolates obtained in this study are in red and extypes taxa are in black bold.
