Colletotrichum truncatum (Schwein.) Andrus & W.D. Moore, Phytopathology 25: 122 (1935). (TNC9) (Ruvi)
MycoBank number: MB 280780; Index Fungorum number: IF 280780; Facesoffungi number: FoF 03827; Fig. **
Saprobic on dead leaves of Dracaena sp. Sexual morph: Undetermined. Asexual morph: Setae abundant, pale to medium brown, smooth walled, 1–5 septate, 100–200 μm (x̅=150) μm (n=10) long, base cylindrical, 4–10 μm (x̅=7) μm (n=20) diam., tip somewhat acute. Conidiophores hyaline, smooth-walled, simple, 10–20 μm (x̅=15) μm (n=20) long. Conidiogenous cells 1–2 μm (x̅=1.5) μm × 2–3.5 μm (x̅=2.75) μm (n=10), hyaline, smooth-walled, cyllindrical to slighty inflated, opening 1–2.5 μm (x̅=1.7) μm (n=20) diam. Collarette present, 0.4–1.1 μm (x̅=0.8) μm (n=20) μm width, periclinal thickening visible. Conidia 20–30 μm (x̅=25) μm × 2–4 μm (x̅=3) μm (n=40), hyaline, smooth or verruculose, aseptate, curved, both sides gradually tapering towards the round to slightly acute apex and truncate base, guttulate.
Material examined – Thailand, Songkhla Province, Hat Yai District, on Dracaena sp., 16 February 2019, Napalai Chaiwan, TNC9 (MFLU 18–****, holotype), ex-type living culture, MFLUCC *****.
Host – chili, dragon fruit, pumpkin tomato and other crops (Chai et al. 2014, Cheng et al. 2014, Diao et al. 2014, Guo et al. 2014). Dracaena sp.—(This study).
Distribution – Australia, China, India, Thailand (Poonpolgul & Kumphai 2007, Than et al. 2008, Sharma et al. 2014, Diao et al. 2015). Thailand—(This study).
GenBank accession numbers – MFLU **; MFLU **; ITS: **; TUB2: ***; ACT: ** CHS ** GAPDH: **
Notes – The first report of Colletotrichum truncatum causes anthracnose diseases of many hosts in bean, soybean, and peanut, chili pepper, pepper and the Solanaceae and Fabaceae (Damm et al. 2009, Shivaprakash et al. 2011). An epitype of this species was described by Damm et al. 2009 as of Phaseolus lunatus in the USA. Damm et al. (2009) synonyms C. capsici and C. curvatum to C. truncatum by used the multilocus phylogenetic analyses. In Australia, most records of this species still require verification by molecular methods (Ford et al. 2004). The closest matches with the sequences in GenBank are all those of the C. truncatum. In a BLASTn search of GenBank, the ITS sequence had ***% similarity (**% nucleotide differences), while the act sequence had **% similarity (** nucleotide differences in 297 nucleotides). Thus, our strain is introduced as a record of C. truncatum from Dracaena sp. in Thailand.

Figure***– Colletotrichum truncatum (MFLU **** TNC9). a, b Fruiting body on dead leaf of Dracaena sp. c–i Conidiophore. j–k Conidiogenous cells l–p Conidia. Scale bars: a = 500 , b = 200 μm, c–i = 20 μm, j–k = 10 μm, l–p = 5 μm.
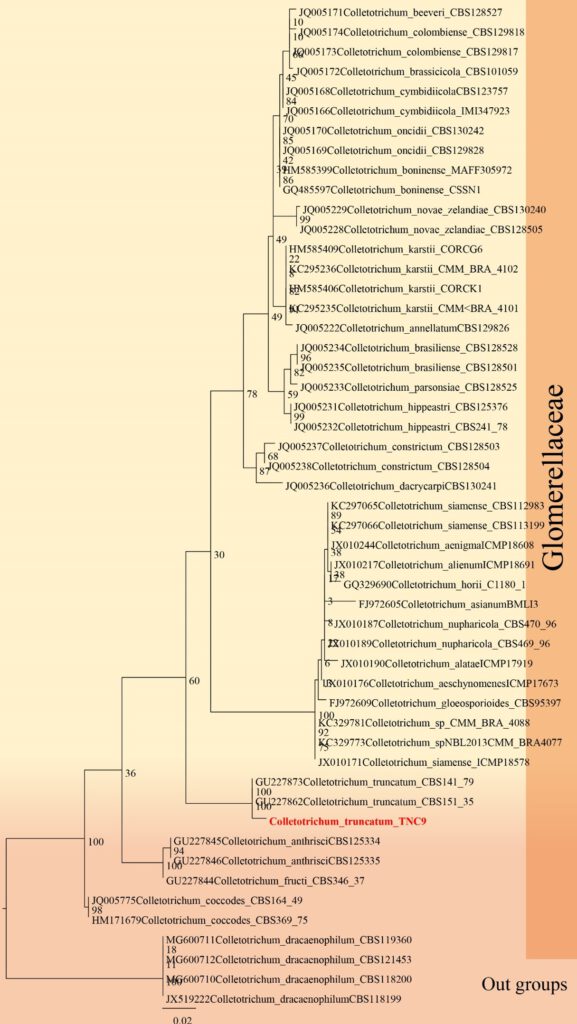
Figure**– Phylogram generated from RAxML analysis based on ITS, CHS, GAPDH and TUB sequence data of selected Glomerellaceae isolates. Related sequences were obtained from GenBank. Twenty-four taxa are included in the analyses, which comprise *** characters including gaps. The tree is rooted to Colletotrichum dracaenophilum. The best scoring RAxML tree with a final likelihood value of − 7281.602128 is presented. The matrix had 613 distinct alignment patterns, with 25.61% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.244440, C = 0.238073, G = 0.310001, T = 0.207486; substitution rates AC = 1.017532, AG = 2.172618, AT = 1.516851, CG = 1.265405, CT = 8.040081, GT = 1.000000; gamma distribution shape parameter α = ***. Maximum likelihood bootstrap support values ≥ 75% (BT) are given at the nodes as ML. The scale bar indicates 0.07 changes. The isolates obtained in this study are in red and extypes taxa are in black bold.
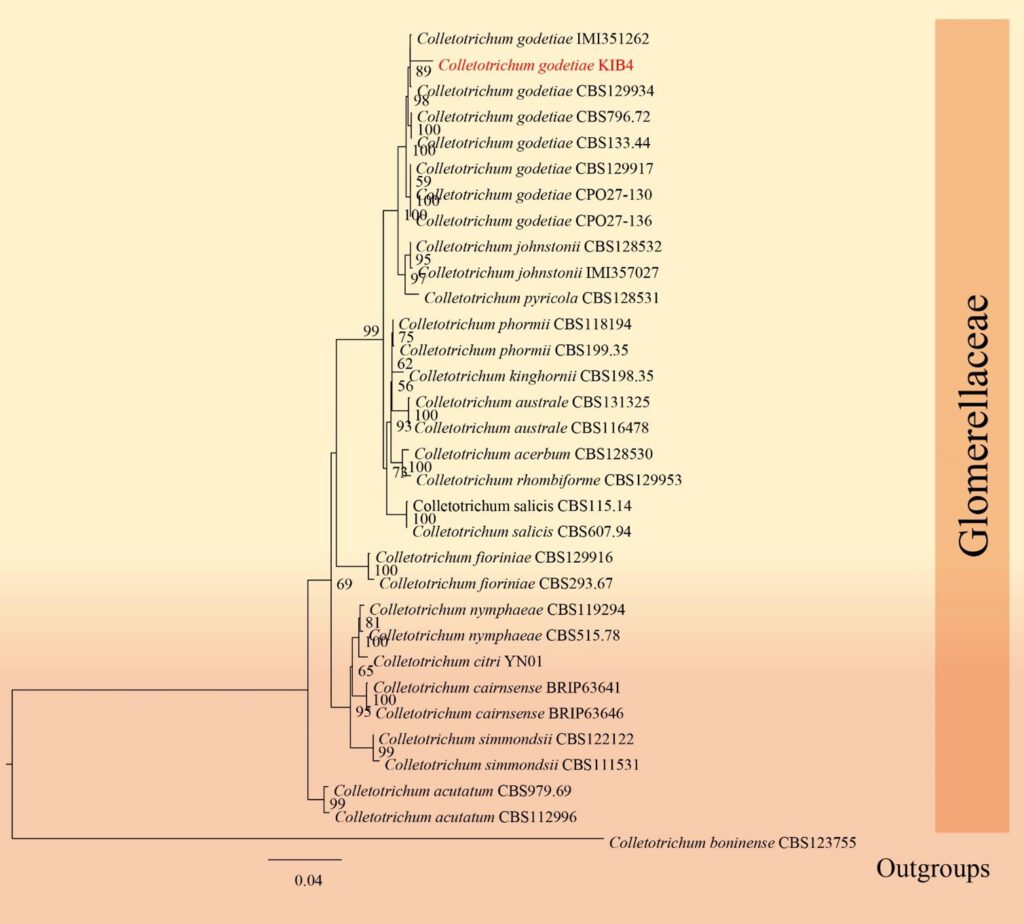
Figure**– Phylogram generated from RAxML analysis based on ITS, GAPDH, CHS, ACT and TUB sequence data of selected Glomerellaceae isolates. Related sequences were obtained from GenBank. Twenty-four taxa are included in the analyses, which comprise 2008 characters including gaps. The tree is rooted to Colletotrichum boninense. The best scoring RAxML tree with a final likelihood value of − 6346.032783 is presented. The matrix had 613 distinct alignment patterns, with 10.47% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.231422, C = 0.297061, G = 0.245294, T = 0.226224; substitution rates AC = 1.502182, AG = 3.422300, AT = 1.013566, CG = 0.595095, CT = 5.885590, GT = 1.000000; gamma distribution shape parameter α = 0.299375. Maximum likelihood bootstrap support values ≥ 75% (BT) are given at the nodes as ML. The scale bar indicates 0.04 changes. The isolates obtained in this study are in red and extypes taxa are in black bold.
