Colletotrichum gigasporum E.F. Rakotoniriana & Munaut, (MRC5) (Ruvi) Mycol. Progr. 12: 407. 2013
= Colletotrichum thailandicum Phouliv., Noireung, L. Cai & K.D. Hyde, Cryptog. Mycol. 33: 354. 2012.
MycoBank number: MB 800175; Index Fungorum number: IF 800175; Facesoffungi number: FoF 10777; Fig. **
Saprobic on dead leaves of Dracaena sp. Sexual morph: Undetermined. Asexual morph: Vegetative hyphae hyaline, smooth-walled, septate, branched. Conidiomata acervular, conidiophores and setae formed on a cushion of roundish to angular brown cells. Setae pale to medium brown, smooth-walled to verruculose 1–3-septate, 50–150 μm long, base cylindrical, 3–5 μm. diam, tip acute to obtuse. Conidiophores pale brown, septate, branched. Conidiogenous cells pale brown, cylindrical to clavate, 15–25 × 4–8 μm, opening 1–2.5 μm diam. Conidia hyaline, aseptate, smooth-walled, cylindrical to slightly curved, both ends rounded, 20–30 × 3.5–6.5 μm.
Material examined – Thailand, Chiang Mai Province, Mae Tang District, on Dracaena sp., 16 February 2019, Napalai Chaiwan, MRC5 (MFLU 18–****, holotype), ex-type living culture, MFLUCC *****.
Host – Centella asiatica, Stylosanthes Guianensis, Coffea arabica (Rakotoniriana et al. 2013). Dracaena sp.—(This study).
Distribution – Colombia, Madagascar, Mexico (Rakotoniriana et al. 2013). Thailand—(This study).
GenBank accession numbers – MFLU **; ITS: **; TUB2: **; ACT: waiting CHS *** GAPDH: ***
Notes – Colletotrichum gigasporum is characterised by large conidia. Phylogenetic analyses based on the ITS and TUB2 sequences placed it in a distinct clade far from the currently accepted Colletotrichum species. Liu et al. 2014 synonym C. thailandicum to C. gigasporum mainly based on ITS and TUB2 sequences from phylogenetic analysis because C. gigasporum was published earlier than C. thailandicum. In addition, C. gigasporum that belongs to the C. gloeosporioides species complex (Jayawardena et al. 2016). Colletotrichum gigasporum can occur from plants as endophytes or pathogens and even strains that were isolated from human tissue and morphology produce aseptate conidia and 0–1-septate ascospores (Rakotoniriana et al. 2013, Tibpromma et al. 2018).
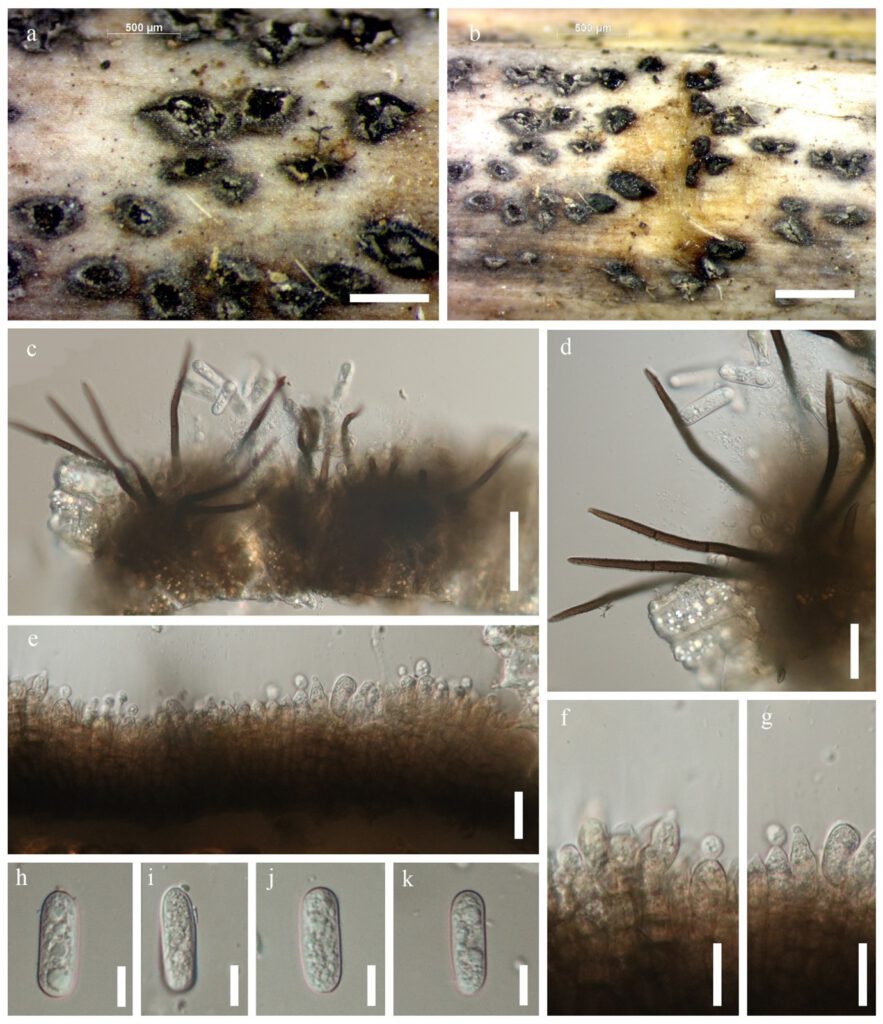
Figure** – Colletotrichum gigasporum (MFLU **** MRC5). a, b Fruiting body on dead leaf of Dracaena sp. c–d Conidiophore. e–g Conidiogenous cells h–k Conidia. Scale bars: a, b = 500 μm, c = 50 μm, d–g = 20 μm, h–k = 10 μm.
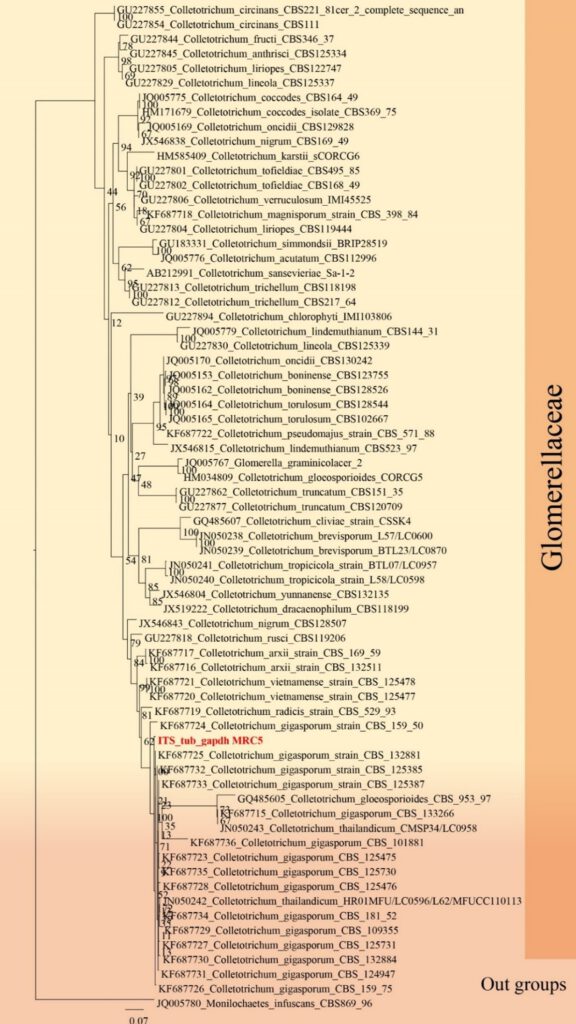
Figure**– Phylogram generated from RAxML analysis based on ITS, TUB and GAPDH sequence data of selected Amplistromataceae isolates. Related sequences were obtained from GenBank. Twenty-four taxa are included in the analyses, which comprise *** characters including gaps. The tree is rooted to Monilochaetes infuscans. The best scoring RAxML tree with a final likelihood value of − 7281.602128 is presented. The matrix had 613 distinct alignment patterns, with 25.61% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.244440, C = 0.238073, G = 0.310001, T = 0.207486; substitution rates AC = 1.017532, AG = 2.172618, AT = 1.516851, CG = 1.265405, CT = 8.040081, GT = 1.000000; gamma distribution shape parameter α = ***. Maximum likelihood bootstrap support values ≥ 75% (BT) are given at the nodes as ML. The scale bar indicates 0.07 changes. The isolates obtained in this study are in red and extypes taxa are in black bold.
