Clonostachys ralfsii Schroers, Studies in Mycology 46, 135 (2001)
Synonyms: Bionectria ralfsii (Berk. & Broome) Schroers & Samuels 2001
Index Fungorum number: IF 485142; Facesoffungi number: FoF 14271; Fig. 1
Saprobic on the dead branch of Corylus avellana. Sexual morph: see (Schroers 2001). Asexual morph: Sporodochium 250–350 μm high, 400–690 at widest sides μm diam. (x̅ = 290 × 550 μm, n = 10), solitary, weakly erumpent, widening in upper part, densely packed subhymenial hyphae or synnematous, yellowish-white patches on the host surface. Conidiophores 3–5.9 μm wide (x̅ = 5.4 μm, n = 20), monomorphic, penicillate, intercalary phialides present, adpressed, in sporodochial aggregates or conidiomata, cylindrical or sub-cylindrical, formed below solitary terminal phialides, apically bearing conidiogenous cells. Conidiogenous cells 5–8 μm wide (x̅ = 6.5 μm, n = 20), enteroblastic, phialidic, arising from a single cell, narrowly flask-shaped, gradually tapering towards the tip with a short collar, conspicuous periclinal thickening, wide near aperture. Conidia 8–13 × 6–8.5 μm (x̅ = 12.1 × 7.5 μm, n = 30), broadly ellipsoidal to limoniform, aseptate, slightly curved, protruding, tapering at both ends, with collarette-like remains of the phialidic tip, deeply pigmented, olive-black conidial mass, hyaline at the beginning, pale green at maturity, finely rough-walled.
Material examined – Italy, Forlì-Cesena Province, Tontola-Predappio, on a dead hanging branch of Corylus avellana, 20 January 2019, Erio Camporesi, IT 4214 (MFLU 19-0611).
GenBank accession numbers – ITS: OP740990, LSU: OP741010.
Known distribution (based on molecular data) – Australia, New Zealand (Schroers 2001), Italy (this study).
Known hosts (based on molecular data) – the bark of unknown dead hosts (Schroers 2001), Corylus avellana (this study).
Notes – In the multi-gene phylogeny (ITS, LSU, β-tubulin and tef1-α) of our study, novel strain (MFLU 19-0611) and other strains of Clonostachys ralfsii (CBS 102845 and CBS 129.87) clustered together with 100% maximum likelihood bootstrap support and 1.00 Bayesian posterior probability in Clade B (Fig. 2). These isolates were collected from unquoted dead hosts from different localities. Detailed sexual and asexual morphology with illustrations of this fungus were provided by Schroers (2001). When comparing the morphology of the asexual morph of C. ralfsii (Schroers 2001), sporodochium, conidiophores, and conidia are slightly larger in our collection (Table 1, Fig. 1). Also, we considered the nucleotide differences within the rRNA gene regions of those strains for further clarification. When comparing the ITS region (ITS1-5.8S-ITS2) of 485 nucleotides, there is one bp (0.20 %) difference in CBS 102845 and CBS 129.87 with our strain (MFLU 19-0611). The LSU rDNA region of MFLU 19-0611 (516 nucleotides) is identical to that of CBS 129.87. Considering the molecular data analysis, we conclude that our new collection is Clonostachys ralfsii.
Table 1 – Synopsis of morphology in asexual morphs of Clonostachys ralfsii
| Character | Schroers (2001) | This study |
|---|---|---|
| Sporodochium | 350–600 µm wide at top | 250–350 μm high, 400–690 at widest sides μm diam. |
| Conidiophores | (2.2–)3.4–4–4.6(–5.4) µm at the widest point | 2.5–5.9 μm wide |
| Conidia | (7.4–)11.6–12.6–13.6(–17.8) ×
(4.8–)6.4–7–7.6(–11) µm |
8–13 μm × 6–8.5 μm |
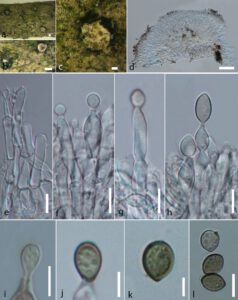
Fig. 1 – Clonostachys ralfsii (MFLU 19-0611, a new host and geographical record). a–c Appearance of sporodochium on the host substrate. d Longitudinal section of a sporodochium. e Appearance of conidiophores in the sporodochium. f–h Different stages of conidiogenesis. i–l Conidia. Scale bars: a, b = 200 μm, c, d = 100 μm, f–h, l = 10 μm, e, i–k = 5 μm.
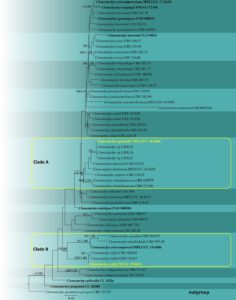
Fig. 2– Phylogram generated from maximum likelihood analysis based on the combined ITS, LSU, β-tubulin and tef1-α sequence data of Clonostachys taxa. Fifty-three strains are included in the combined analyses which comprised 2376 characters (493 characters for ITS, 850 characters for LSU, 648 characters for β-tubulin, 385 characters for tef1-α) after aligned. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RAxML tree with a final likelihood value of -10039.778526 is presented. The matrix had 632 distinct alignment patterns, with 51.62% undetermined characters or gaps. Evolutionary models applied for ITS, LSU, β-tubulin and tef1-α genes are SYM+I+G, GTR+I+G, HKY+I+G and K80+I models, respectively. Bootstrap support values for ML equal to or greater than 70% and Bayesian posterior probabilities equal to or greater than 0.95 are given near nodes, respectively. The tree is rooted with Clonostachys grammicosporopsis (CBS 115.87). Ex-type strains are in bold. The newly generated sequences are indicated in yellow.
