Clonostachys agarwalii (Kushwaha) Schroers [as ‘agrawalii’], Studies in Mycology 46, 90 (2001)
Basionym: Gliocladium agarwalii Kushwaha, Current Science 49(2), 74 (1980)
Index Fungorum number: IF 485123; Facesoffungi number: FoF 14270; Fig. 1
Saprobic on dead twigs of Lagerstroemia sp. Sexual morph: Undetermined. Asexual morph: Conidiomata sporodochial. Sporodochia somewhat synnematous, short stipitate, scattered or arranged in small groups on the substrate. Stipe up to 145 μm tall, 95–250 μm wide. Conidiophores 23–30 × 2–2.5 μm (x̄ = 27 × 2.2 μm, n = 15), monomorphic, penicillate, mononematous, densely aggregated, hyaline, septate, short stipitate. Conidiogenous cells 10–15 × 2.1–3.3 μm (x̄ = 12.7 × 2.8 μm, n = 25), phialidic, arranged in groups of 2 at the tip of conidiophores, narrowly flask-shaped, hyaline, straight to slightly curved, smooth-walled, periclinal thickening at the tip, with a visible collarette. Conidia 3.5–5.1 × 2.2–2.7 μm (x̄ = 4 × 2.5 μm, n = 55), ovoidal to ellipsoidal, somewhat curved or straight, mostly with a slightly flattened side, aseptate, hilum laterally displaced, arranged in imbricate white to pale orange columns, can breakdown into slimy masses, smooth-walled.
Material examined – Thailand, Chiang Mai Province, Mae Rim Sub-district, dead twigs of Lagerstroemia sp., 18 September 2017, RH Perera, Rim15 (MFLU 19-0976), living culture MFLUCC 18-0568.
GenBank accession numbers – ITS: OM276726, LSU: OM276821, tef1-α: OM283276.
Known distribution (based on molecular data) – India (Schroers 2001), Thailand (this study).
Known hosts (based on molecular data) – decomposing buffalo horn pieces (Schroers 2001), Lagerstroemia sp. (this study).
Notes – Our new isolate (MFLUCC 18-0568) grouped with Clonostachys agarwalii (CBS 533.81) and three endophytic Clonostachys isolates (DJL25, DJL21, and DJL16) obtained from root samples of Changnienia amoena (Orchidaceae) in China (Fig. 2). No morphological data have been provided for those endophyte isolates (Jiang et al. 2011). Our collection (MFLU 19-0976) fits the description of conidiogenous cells and conidial measurements of C. agarwalii provided by Schroers (2001). With the evidence from the phylogenetic analysis and morphology, we identify our isolate as C. agarwalii. We collected sporodochia from the natural substrate, but none were reported from in vitro cultures (Schroers 2001). This paper illustrates Clonostachys agarwalii as a new host of Lagerstroemia sp. and a new geographical record from Thailand.
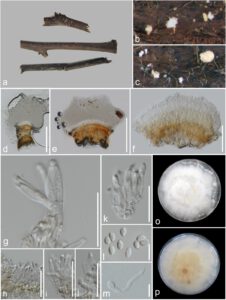
Fig. 1 – Clonostachys agarwalii (MFLU 19-0976, new host and geographical record). a Herbarium material. b, c Sporodochia on the substrate. d–f Sporodochia mounted in water. g–k Conidiophores with phialides and conidia. l Conidia. m Germinating conidium. o, p Colony on PDA. Scale bars: d, e = 200 µm, f = 50 µm, g–i = 20 µm, j, k = 10 µm.
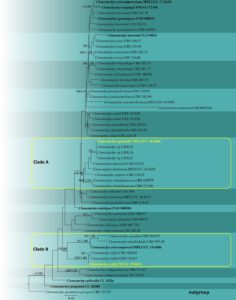
Fig. 2 – Phylogram generated from maximum likelihood analysis based on the combined ITS, LSU, β-tubulin and tef1-α sequence data of Clonostachys taxa. Fifty-three strains are included in the combined analyses which comprised 2376 characters (493 characters for ITS, 850 characters for LSU, 648 characters for β-tubulin, 385 characters for tef1-α) after aligned. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RAxML tree with a final likelihood value of -10039.778526 is presented. The matrix had 632 distinct alignment patterns, with 51.62% undetermined characters or gaps. Evolutionary models applied for ITS, LSU, β-tubulin and tef1-α genes are SYM+I+G, GTR+I+G, HKY+I+G and K80+I models, respectively. Bootstrap support values for ML equal to or greater than 70% and Bayesian posterior probabilities equal to or greater than 0.95 are given near nodes, respectively. The tree is rooted with Clonostachys grammicosporopsis (CBS 115.87). Ex-type strains are in bold. The newly generated sequences are indicated in yellow.
