Boothiella Lodhi & Mirza, Mycologia 54(2): 217 (1962)
MycoBank number: MB 627; Index Fungorum number: IF 627, Facesoffungi number: FoF14655
Isolated from soil. Sexual morph: Ascomata cleistothecial, solitary or gregarious, superficial to immersed in the medium, globose to subglobose, membranaceous, brown to black, glabrous. Peridium composed of membranaceous, hyaline to subhayline cells of textura angularis. Asci 4-spored, unitunicate, clavate to cylindrical, short pedicellate, apex rounded, evanescent. Ascospores hyaline, becoming dark brown, oval to citriform, 1–2-apiculate, aseptate, with a germ pore at one or each end. Asexual morph: Undetermined (adapted from Lodhi and Mirza 1962; Wang et al. 2019a, b).
Notes – The monotypic genus Boothiella was introduced as a member of Eurotiales and its ascospores are similar to Thielavia (Lodhi and Mirza 1962). Kirk et al. (2008) transferred Boothiella to Sordariaceae and Maharachchikumbura et al. (2015, 2016) placed this genus in Chaetomiaceae. Vu et al. (2019) and Wang et al. (2019a, b) analyzed sequence data from isolates CBS 334.67 (ex-type) and CBS 887.97, and showed that B. tetraspora nests in Sordariaceae. In the present study, Boothiella strains form a clade in Sordariaceae (100%ML/1.00PP, Figs. 26, 40).
Type species – Boothiella tetraspora Lodhi & J.H. Mirza, Mycologia 54(2): 217 (1962)
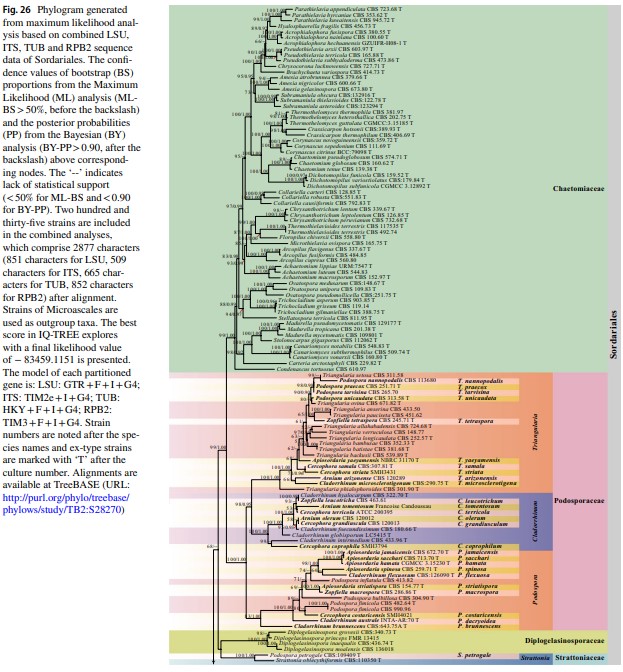
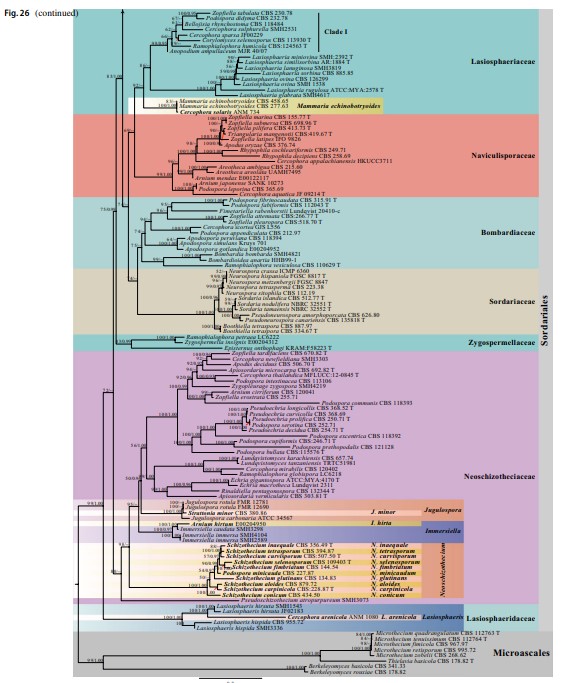

Fig. 40 Phylogram generated from maximum likelihood analysis based on combined LSU, ITS and TUB sequence data of Sordari- aceae. The confidence values of bootstrap (BS) proportions from the Maximum Likelihood (ML) analysis (ML-BS > 50%, before the back- slash) and the posterior probabilities (PP) from the Bayesian (BY) analysis (BY-PP > 0.90, after the backslash) above corresponding nodes. The ‘–’ indicates lack of statistical support (< 50% for ML-BS and < 0.90 for BY-PP). Related sequences were referred to Cai et al. (2006b), Wang et al. (2019a, b) and Vu et al. (2019). Eighty-four strains are included in the combined analyses which comprise 1909 characters (862 characters for LSU, 583 characters for ITS, 464 characters for TUB) after alignment. Strains of Cephalothecales are used as the outgroup taxa. The model of each partitioned gene is GTR + I + G. The best score in IQ-TREE explores with a final like- lihood value of -9569.3117 is presented. Sequences generated indi- cated in bold. Strain numbers are noted after the species names and ex-type strains are marked with ‘T’ after the culture number. Some important characters of Sordariaceae species treated in this study are illustrated at the right side of the tree. Alignments are available at TreeBASE (URL: http://purl.org/phylo/treebase/phylows/study/TB2: S28269)
Species
