Bacusphaeria Norlailatul, Alias et Suetrong, Botanica Marina 60 (4): 11 (2017)
MycoBank number: MB 812182; Index Fungorum number: IF 812182; Facesoffungi number: FoF 04318; 1 species with sequence data.
Type species – Bacusphaeria nypae Norlailatul, Alias & Suetrong
Notes – The monotypic genus Bacusphaeria is similar to Tirisporella, because they have large, thick-walled ascomata, cylindrical asci and both occur on the basal petioles of Nypa fruticans in brackish water. However, Bacusphaeria is distinct from Tirisporella because it; i) lacks complex ascospore appendages; ii) has fewer septa in the ascospores (Bacusphaeria: 1–5-septate vs. Tirisporella: 5–7-septate); iii) lack of paraphyses; iv) has a prominent apical ring in contrast to faint apical ring in Tirisporella; v) having generally uniseriate ascospores in contrast to biseriate ascospores in Tirisporella. Base pair differences of Bacusphaeria nypae (MFLU 13-0617) and Tirisporella beccariana (BCC38300) are 97 bp different out of 859 bp (11.29%) without gaps in the LSU gene locus. Furthermore, in phylogeny, Bacusphaeria nypae is distinct from the other two genera and formed a separate lineage in Tirisporellaceae (Fig. 7).
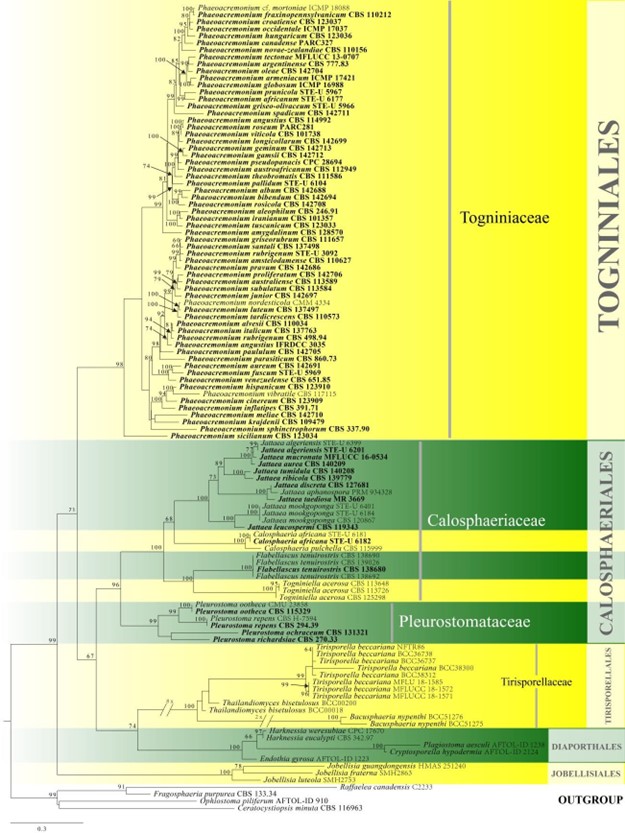
Figure 7 – Phylogram generated from maximum likelihood analysis based on combined tub2, act, ITS and LSU sequence data of Calosphaeriales, Togniniales, Jobellisiales and Tirisporallales. One hundred and fifteen strains are included in the combined analyses which comprised 2296 characters (572 characters for tub2, 269 characters for act, 586 characters for ITS, 869 characters for LSU) after alignment. Three species of Ophiostomataceae are used as outgroup taxa. Single gene
analyses were also carried out and the phylogenies were similar in topology and clade stability. The 377 best RAxML tree with a final likelihood value of -29642.219830 is presented. Estimated base frequencies were as follows: A = 0.222849, C = 0.289953, G = 0.265413, T = 0.221786; substitution rates AC = 1.361161, AG = 3.471108, AT = 1.335451, CG = 1.139165, CT = 4.985065, GT = 1.000000; gamma distribution shape parameter a = 1.412987. Bootstrap support
values for ML greater than 60% are given near the nodes. Ex-type strains are in bold. The newly generated sequences are indicated in blue
Species
