Acrocalymma pterocarpi Jayasiri, E.B.G. Jones & K.D. Hyde, in Jayasiri et al., Mycosphere 10(1), 20 (2019)
Index Fungorum number: IF 555528; FacesofFungi number: FoF 05228; Fig. 1
Saprobic on dead twigs of Bidens sp. Sexual morph: Ascomata 120–130 μm high, 110–130 μm diam. (x̄ = 125 × 120 μm, n = 10), scattered, erumpent to nearly superficial, globose to subglobose, dark brown to black, ostiolate with minute papilla. Ostiole 34–36 μm diam., central, papillate. Peridium 10–30 μm diam., composed of several layers of small, flattened, brown to dark brown pseudoparenchymatous cells, inner cells hyaline, arranged in a textura angularis. Hamathecium composed of 1–2 μm wide, numerous, filamentous, branched, septate, pseudoparaphyses. Asci 40–60 × 8–10 μm (x̄ = 50 × 9.5 μm, n = 20), 8-spored, bitunicate, fissitunicate, cylindrical, with a short, narrowed, with furcate pedicel, apically rounded with a small ocular chamber. Ascospores 16–20 × 5–6 μm (x̄ = 19 × 5.5 μm, n = 30), overlapping biseriate, hyaline, fusiform, 1–3-septate, guttulate, with narrowly rounded ends with mucilaginous sheath. Asexual morph: Undetermined.
Material examined – Thailand, Chiang Mai Province, Mushroom Research Centre (MRC), on dead twig of Bidens sp., 11 November 2020, Pahoua Pasouvang, MFLU 23-0002.
GenBank Number – ITS: OQ184951, LSU: OQ184954, SSU: OQ184984
Known distribution (based on molecular data) – China (de Silva et al. 2022), Thailand (Jayasiri et al. 2019, this study).
Known hosts (based on molecular data) – On a fallen pod of Pterocarpus indicus (Jayasiri et al. 2019), dead twigs attached to the Magnolia sp. (de Silva et al. 2022), dead twigs of Bidens sp. (this study).
Notes – Morphologically, our collection (MFLU 23-0002) is similar to the holotype of A. pterocarpi (MFLUCC 17–0926) collected from a fallen pod of Pterocarpus indicus (Fabaceae) in Thailand (Jayasiri et al. 2019). Both isolates have similar morphology (Fig. 1), with erumpent to nearly superficial, globose or subglobose ascomata, cylindrical asci with a short, narrowed, furcate pedicel and hyaline, fusiform, 1–3-septate ascospores (Jayasiri et al. 2019). However, the size of ascomata (x̄ = 125 × 120 μm) and asci (x̄ = 50 × 9.5 μm) of our collection are comparatively smaller than the holotype (x̄ = 143 × 141 μm vs. x̄ = 70 × 10 μm) (Jayasiri et al. 2019). According to the phylogenetic analyses (Fig. 2), our collections (MFLU 23-0002) clustered with A. pterocarpi strain (MFLUCC 18-0718) with 97% maximum parsimony bootstrap support and 0.99 Bayesian posterior probability. Furthermore, base pair differences between our collection (MFLUCC 17-0926) and the holotype of A. pterocarpi (MFLUCC 17-0926) are insignificant (LSU = 0.4% (3/760 bp), SSU = 0.4% (4/985 bp), and ITS = 1.2% (6/468 bp), confirming that they are the same species. Based on the morpho-molecular evidence, we introduced our new collection as a new host record of A. pterocarpi from Bidens sp. in Thailand.
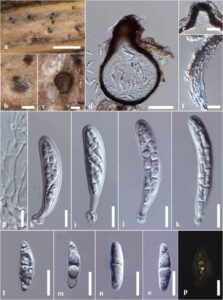
Fig. 1 – Acrocalymma pterocarpi on a dead twig of Bidens sp. (MFLU 23-0002, a new host record). a, b Appearance of ascomata on host surface. c, d Section through an ascoma. e Ostiole. f Section through the peridium. g Pseudoparaphyses. h–k Asci. l–o Ascospores. p An ascospore stained with Indian Ink. Scale bars: a =500 μm, b =100 μm, c, d =50 μm, e =20 μm, f–p =10 μm.
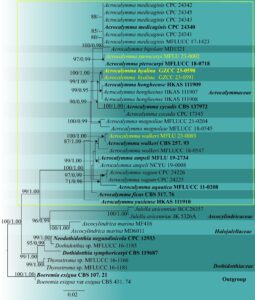
Fig. 2 – Phylogram generated from the maximum likelihood analysis based on the combined LSU, SSU and ITS sequence data. Thirty-nine strains are included in the combined analyses. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RAxML tree with a final likelihood value of -6868.654138 is presented. Evolutionary models applied for all genes are GTR+G. The matrix had 483 distinct alignment patterns, with 28.24% of undetermined characters or gaps. Bootstrap support values for maximum likelihood greater than 70% and Bayesian posterior probabilities greater than 0.95 are given near nodes, respectively. The tree was rooted with Boeremia exigua (CBS 107. 21) and B. exigua var exigua (CBS 431. 74). Ex-type strains are in bold. The newly generated sequences are indicated in yellow.
