Xenovaginatispora phichaiensis Boonmee, Huanraluek & K.D. Hyde, sp. nov.
MycoBank number: MB 558537; Index Fungorum number: IF 558537; Facesoffungi number: FoF 09191; Fig. 36
Etymology: The specific epithet “phichaiensis” refers to the Phichai District, Uttaradit Province, Thailand.
Holotype: MFLU 21-0059
Saprobic on submerged decaying wood in a freshwater stream. Sexual morph Ascomata 151–225 µm high, 187–211 µm diam., immersed to semi-immersed, erumpent, solitary, scattered, globose to subglobose, dark brown, coriaceous to carbonaceous, ostiolate. Papilla slit-like, variable in shape, central papilla, coriaceous, crest-like at the apex, with an irregular pore-like opening. Peridium 27–38 µm wide, composed of several layers of dark brown cells of textura angularis, merged with the host tissues. Hamathecium comprising numerous, 1.5–3.5 µm wide, filamentous, rarely branched and anastomosed, hyaline, pseudoparaphyses, embedded in a gelatinous matrix. Asci 98–115.5 × 22–28 µm (x̅ = 107 × 24 µm, n = 15), 8-spored, bitunicate, elongate ovoid to clavate, short pedicellate, with knob-like or obtuse, slightly narrow to rounded at the apex. Ascospores 35–42 × 10–13 µm (x̅ = 39 × 12 µm, n = 10), overlapping 1–2-seriate, broadly fusoid to fusiform, 1-septate, slightly constricted at the septum, both cells at central septum wider, with a single large guttule, 8–12 µm diam., towards the centre of each cell, surrounded by a prominent and thick-walled mucilaginous sheath with wavy edge, drawn out towards each end to form tapering appendages. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on PDA within 24 h and germ tubes arising from both ends. Colonies on MEA, slow growing, reaching 3 cm diam. after 1 month of incubation at room temperature, initially dull white to light grey becoming dark grey at maturity, slightly effuse, radially with an undulate edge, greyish, light brown with brown pigmented in media, no sporulating in culture after 60 days.
Material examined: THAILAND, Uttaradit Province, Phichai, Thasak, Chom Tok Village, on submerged decaying wood in a small freshwater stream, 29 December 2018, S. Boonmee, CTU8 (MFLU 21-0059, holotype), ex-type living culture, MFLUCC 21-0082.
GenBank numbers: ITS = MZ538534, LSU = MZ538569, SSU = MZ538577, TEF1-α = MZ567110.
Notes: In the NCBI BLASTn search of LSU and SSU sequences, Xenovaginatispora phichaiensis MFLUCC 21-0082 is most similar to an undescribed taxon Pleosporales sp. (F65-1) with 99.19% (LSU) and 99.70% (SSU) similarity. Unfortunately, we cannot compare the morphology in Shearer et al. (2009) as it lacks a description. Multi-gene phylogenetic analysis indicates that Xenovaginatispora phichaiensis clusters with the undescribed Pleosporales sp. (F65-1) and both of them form a subclade basal to Neolindgomyces species (Lindgomycetaceae) with 93% MLBS, 1.00 BYPP support (Fig. 37). Xenovaginatispora phichaiensis differs from Neolindgomyces species (Jayasiri et al. 2019) in having characteristics such as elongate ovoid to clavate asci and broadly fusoid to fusiform, only 1-septate ascospores, with two distinct large guttules in both cells. Therefore, we introduce X. phichaiensis as a new species based on morphology and phylogenetic evidences (Figs. 36 and 37).
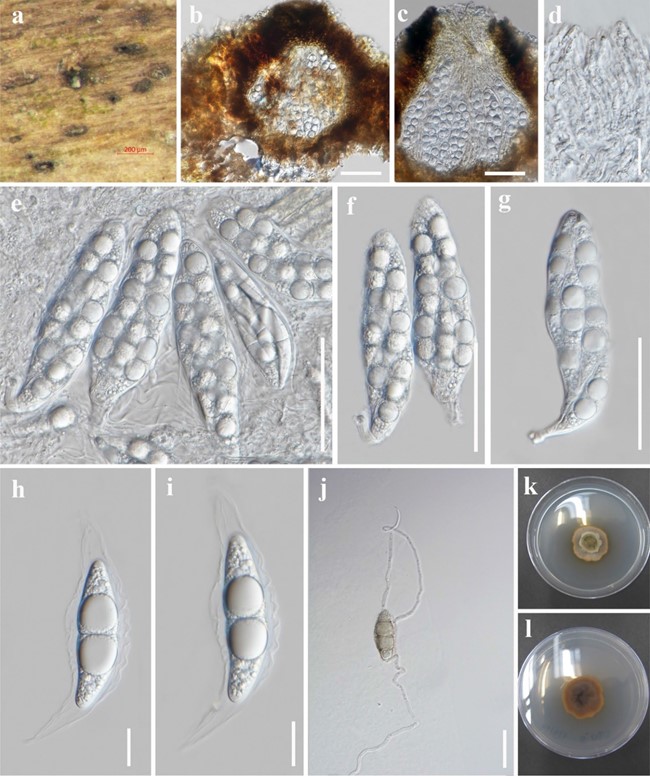
Fig. 36 Xenovaginatispora phichaiensis (MFLU 21-0059, holo- type). a Appearance of ascomata. b Longitudinal sections of ascomata. c Ascomata with ostiole. d Pseudoparaphyses. e–g Asci. h, i Ascospores surrounded by mucilaginous sheath and distinct api- cal appendages. j Germinated ascospore. k, l Culture on MEA at 1 month (k = from surface and l = reverse). Scale bars: b = 100 μm, c, d = 50 μm, e–g, j = 20 μm, h–i = 10 μm
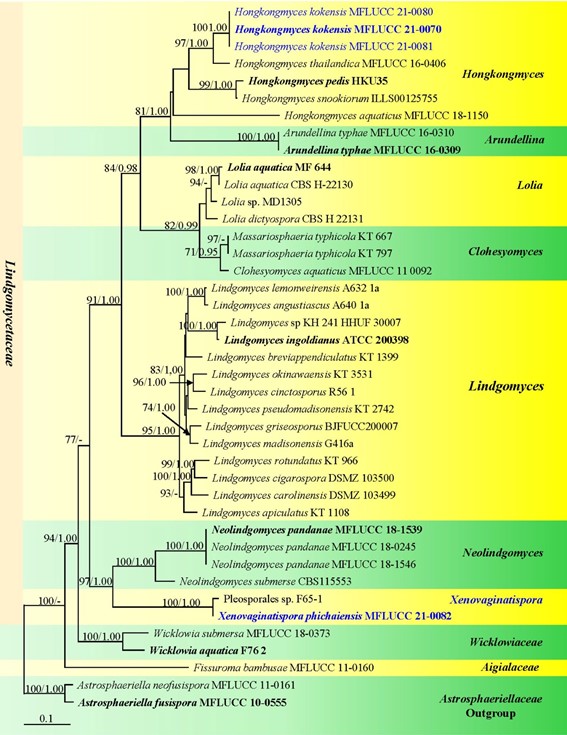
Fig. 37 Phylogram generated from maximum likelihood analysis based on combined LSU, SSU, ITS, and TEF1-α sequence data of new species and new genus in Lindgomycetaceae. Astrosphaeriella neofusispora MFLUCC 11-0161 and A. fusispora MFLUCC 10-0555 are selected as the outgroup taxa. Bootstrap support values for maxi-mum likelihood (MLBS, left) equal to or greater than 70% is given above the nodes. Bayesian posterior probabilities (BYPP, right) equal to or greater than 0.95 are given above the nodes. Ex-type strains are in bold and newly generated sequences are in blue
